A novel approach to dissect the abscission process in Arabidopsis
- PMID: 22992509
- PMCID: PMC3490599
- DOI: 10.1104/pp.112.205955
A novel approach to dissect the abscission process in Arabidopsis
Abstract
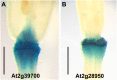
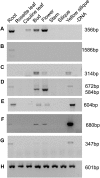
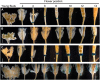
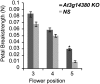
Abscission is the consequence of a specialized layer of cells undergoing a complex series of molecular and biochemical events. Analysis of the specific molecular changes associated with abscission is hampered by contamination from neighboring nonseparating tissues. Moreover, studies of abscission frequently involve the examination of events that take place in isolated segments of tissue exposed to nonphysiological concentrations of ethylene or indole-3-acetic acid for protracted periods (more than 24 h) of time. To resolve these problems, we have adopted the use of a transgenic line of Arabidopsis (Arabidopsis thaliana) where the promoter of an abscission-specific polygalacturonase gene (At2g41850/ARABIDOPSIS DEHISCENCE ZONE POLYGALACTURONASE2) has been fused to a green fluorescent protein reporter. RNA was extracted from green fluorescent protein-tagged cells, released from abscising floral organs, and used to generate a complementary DNA library. This library was used to probe a microarray, and a population of abscission-related transcripts was studied in detail. Seven novel abscission-related genes were identified, four of which encode proteins of unknown function. Reverse transcription-polymerase chain reaction analyses and promoter fusions to the β-glucuronidase reporter gene confirmed the expression of these genes in the abscission zone and revealed other places of expression during seedling development. Three of these genes were studied further by crossing reporter lines to the abscission mutants inflorescence deficient in abscission (ida) and blade-on-petiole1 (bop1)/bop2 and an IDA-overexpressing line. Phenotypic analysis of an At3g14380 transfer DNA insertion line indicates that this gene plays a functional role in floral organ shedding. This strategy has enabled us to uncover new genes involved in abscission, and their possible contribution to the process is discussed.
Figures




References
-
- Adamczyk BJ, Lehti-Shiu MD, Fernandez DE. (2007) The MADS domain factors AGL15 and AGL18 act redundantly as repressors of the floral transition in Arabidopsis. Plant J 50: 1007–1019 - PubMed
-
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al. (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 - PubMed
-
- Belfield EJ, Ruperti B, Roberts JA, McQueen-Mason S. (2005) Changes in expansin activity and gene expression during ethylene-promoted leaflet abscission in Sambucus nigra. J Exp Bot 56: 817–823 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

