Genomic analysis of a key innovation in an experimental Escherichia coli population
- PMID: 22992527
- PMCID: PMC3461117
- DOI: 10.1038/nature11514
Genomic analysis of a key innovation in an experimental Escherichia coli population
Abstract
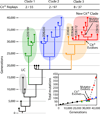
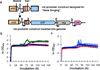
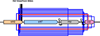
Evolutionary novelties have been important in the history of life, but their origins are usually difficult to examine in detail. We previously described the evolution of a novel trait, aerobic citrate utilization (Cit(+)), in an experimental population of Escherichia coli. Here we analyse genome sequences to investigate the history and genetic basis of this trait. At least three distinct clades coexisted for more than 10,000 generations before its emergence. The Cit(+) trait originated in one clade by a tandem duplication that captured an aerobically expressed promoter for the expression of a previously silent citrate transporter. The clades varied in their propensity to evolve this novel trait, although genotypes able to do so existed in all three clades, implying that multiple potentiating mutations arose during the population's history. Our findings illustrate the importance of promoter capture and altered gene regulation in mediating the exaptation events that often underlie evolutionary innovations.
Figures

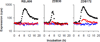


Comment in
-
Evolution: How the unicorn got its horn.Nature. 2012 Sep 27;489(7417):504-5. doi: 10.1038/nature11487. Epub 2012 Sep 19. Nature. 2012. PMID: 22992522 No abstract available.
-
Bacterial evolution: decoding the bacterial fossil record.Nat Rev Microbiol. 2012 Nov;10(11):729. doi: 10.1038/nrmicro2895. Epub 2012 Oct 1. Nat Rev Microbiol. 2012. PMID: 23023737 No abstract available.
-
Bacterial evolution: Decoding fossil records.Nat Rev Genet. 2012 Nov;13(11):757. doi: 10.1038/nrg3348. Epub 2012 Oct 3. Nat Rev Genet. 2012. PMID: 23032254 No abstract available.
References
-
- Mayr E. The emergence of evolutionary novelties. In: Tax S, editor. Evolution after Darwin. Chicago: University of Chicago Press; 1960.
-
- Pigliucci M. What, if anything, is an evolutionary novelty? Philos. Science. 2008;75:887–898.
-
- Jacob F. Evolution and tinkering. Science. 1977;196:1161–1166. - PubMed
-
- Jacob F. The Possible and the Actual. Seattle: Univ. Washington Press; 1982.
-
- Gould SJ, Vrba ES. Exaptation – a missing term in the science of form. Paleobiol. 1982;8:4–15.

