Assembly of allosteric macromolecular switches: lessons from PKA
- PMID: 22992589
- PMCID: PMC3985763
- DOI: 10.1038/nrm3432
Assembly of allosteric macromolecular switches: lessons from PKA
Abstract
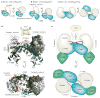
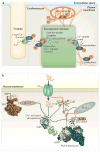
Protein kinases are dynamic molecular switches that have evolved to be only transiently activated. Kinase activity is embedded within a conserved kinase core, which is typically regulated by associated domains, linkers and interacting proteins. Moreover, protein kinases are often tethered to large macromolecular complexes to provide tighter spatiotemporal control. Thus, structural characterization of kinase domains alone is insufficient to explain protein kinase function and regulation in vivo. Recent progress in structural characterization of cyclic AMP-dependent protein kinase (PKA) exemplifies how our knowledge of kinase signalling has evolved by shifting the focus of structural studies from single kinase subunits to macromolecular complexes.
Conflict of interest statement
The authors declare no competing financial interests.
Figures




References
-
- Krebs EG, Graves DJ, Fischer EH. Factors affecting the activity of muscle phosphorylase B kinase. J Biol Chem. 1959;234:2867–2873. - PubMed
-
- Pidoux G, Tasken K. Specificity and spatial dynamics of protein kinase A signaling organized by A-kinase-anchoring proteins. J Mol Endocrinol. 2010;44:271–284. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

