Insights into avian influenza virus pathogenicity: the hemagglutinin precursor HA0 of subtype H16 has an alpha-helix structure in its cleavage site with inefficient HA1/HA2 cleavage
- PMID: 22993148
- PMCID: PMC3497694
- DOI: 10.1128/JVI.01606-12
Insights into avian influenza virus pathogenicity: the hemagglutinin precursor HA0 of subtype H16 has an alpha-helix structure in its cleavage site with inefficient HA1/HA2 cleavage
Abstract
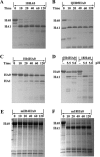
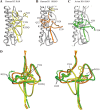
With a new serotype (H17) of hemagglutinin (HA) recently being discovered, there are now 17 serotypes (H1 to H17) of influenza A viruses in total. It is believed that HA is initially expressed as a precursor of HA0 and then cleaved into HA1 and HA2, forming a disulfide bond-linked complex, for its full function. Structural data show that a loop structure exists in the cleavage site between HA1 and HA2, and this flexible loop is crucial for the efficient cleavage of HA0. Here, the crystal structures of H16 (a low-pathogenicity avian influenza virus) in their HA0 form (H16HA0) have been solved at 1.7-Å and 2.0-Å resolutions. To our surprise, an α-helix element in the cleavage site which inserts into the negatively charged cavity with the key residue R329 hidden behind the helix was observed. In vitro trypsin cleavage experiments demonstrated inefficient cleavage of H16HA0 under both neutral and low-pH conditions. The results provide new insights into influenza A virus pathogenicity; both the relatively stable α-helix structure in the flexible cleavage loop and inaccessibility of the cleavage site likely contribute to the low pathogenicity of avian influenza A virus. Furthermore, compared to all of the HAs whose structures have been solved, H16 is a good reference for assigning the HA subtypes into two groups on the basis of the three-dimensional structure, which is consistent with the phylogenetic grouping. We conclude that in light of the current H16HA0 structure, the natural α-helix element might provide a new opportunity for influenza virus inhibitor design.
Figures




References
-
- Anonymous 1994. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 50:760–763 - PubMed
-
- Bullough PA, Hughson FM, Skehel JJ, Wiley DC. 1994. Structure of influenza haemagglutinin at the pH of membrane fusion. Nature 371:37–43 - PubMed
-
- Chen J, et al. 1998. Structure of the hemagglutinin precursor cleavage site, a determinant of influenza pathogenicity and the origin of the labile conformation. Cell 95:409–417 - PubMed
-
- Corti D, et al. 2011. A neutralizing antibody selected from plasma cells that binds to group 1 and group 2 influenza A hemagglutinins. Science 333:850–856 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

