A network of protein interactions around the herpes simplex virus tegument protein VP22
- PMID: 22993164
- PMCID: PMC3497626
- DOI: 10.1128/JVI.01913-12
A network of protein interactions around the herpes simplex virus tegument protein VP22
Abstract
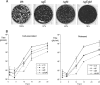
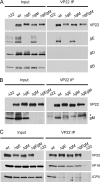
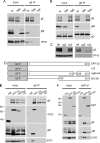
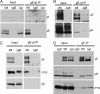
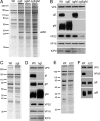
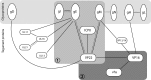
Assembly of the herpesvirus tegument is poorly understood but is believed to involve interactions between outer tegument proteins and the cytoplasmic domains of envelope glycoproteins. Here, we present the detailed characterization of a multicomponent glycoprotein-tegument complex found in herpes simplex virus 1 (HSV-1)-infected cells. We demonstrate that the tegument protein VP22 bridges a complex between glycoprotein E (gE) and glycoprotein M (gM). Glycoprotein I (gI), the known binding partner of gE, is also recruited into this gE-VP22-gM complex but is not required for its formation. Exclusion of the glycoproteins gB and gD and VP22's major binding partner VP16 demonstrates that recruitment of virion components into this complex is highly selective. The immediate-early protein ICP0, which requires VP22 for packaging into the virion, is also assembled into this gE-VP22-gM-gI complex in a VP22-dependent fashion. Although subcomplexes containing VP22 and ICP0 can be formed when either gE or gM are absent, optimal complex formation requires both glycoproteins. Furthermore, and in line with complex formation, neither of these glycoproteins is individually required for VP22 or ICP0 packaging into the virion, but deletion of gE and gM greatly reduces assembly of both VP22 and ICP0. Double deletion of gE and gM also results in small plaque size, reduced virus yield, and defective secondary envelopment, similar to the phenotype previously shown for pseudorabies virus. Hence, we suggest that optimal gE-VP22-gM-gI-ICP0 complex formation correlates with efficient virus morphogenesis and spread. These data give novel insights into the poorly understood process of tegument acquisition.
Figures


Similar articles
-
Virion incorporation of the herpes simplex virus type 1 tegument protein VP22 occurs via glycoprotein E-specific recruitment to the late secretory pathway.J Virol. 2009 May;83(10):5204-18. doi: 10.1128/JVI.00069-09. Epub 2009 Mar 11. J Virol. 2009. PMID: 19279114 Free PMC article.
-
Cytoplasmic residues of herpes simplex virus glycoprotein gE required for secondary envelopment and binding of tegument proteins VP22 and UL11 to gE and gD.J Virol. 2007 Jan;81(1):319-31. doi: 10.1128/JVI.01842-06. Epub 2006 Oct 11. J Virol. 2007. PMID: 17035313 Free PMC article.
-
A conserved region of the herpes simplex virus type 1 tegument protein VP22 facilitates interaction with the cytoplasmic tail of glycoprotein E (gE).Virology. 2007 Feb 5;358(1):192-200. doi: 10.1016/j.virol.2006.08.024. Epub 2006 Sep 25. Virology. 2007. PMID: 16997344
-
Recruitment of herpes simplex virus type 1 immediate-early protein ICP0 to the virus particle.J Virol. 2010 May;84(9):4682-96. doi: 10.1128/JVI.00126-10. Epub 2010 Feb 17. J Virol. 2010. PMID: 20164220 Free PMC article.
-
Tegument Assembly and Secondary Envelopment of Alphaherpesviruses.Viruses. 2015 Sep 18;7(9):5084-114. doi: 10.3390/v7092861. Viruses. 2015. PMID: 26393641 Free PMC article. Review.
Cited by
-
HSV-1 gM and the gK/pUL20 complex are important for the localization of gD and gH/L to viral assembly sites.Viruses. 2015 Mar 4;7(3):915-38. doi: 10.3390/v7030915. Viruses. 2015. PMID: 25746217 Free PMC article.
-
Functional interactions between herpes simplex virus pUL51, pUL7 and gE reveal cell-specific mechanisms for epithelial cell-to-cell spread.Virology. 2019 Nov;537:84-96. doi: 10.1016/j.virol.2019.08.014. Epub 2019 Aug 18. Virology. 2019. PMID: 31493658 Free PMC article.
-
Bovine Herpesvirus 1 UL49.5 Interacts with gM and VP22 To Ensure Virus Cell-to-Cell Spread and Virion Incorporation: Novel Role for VP22 in gM-Independent UL49.5 Virion Incorporation.J Virol. 2018 Jun 13;92(13):e00240-18. doi: 10.1128/JVI.00240-18. Print 2018 Jul 1. J Virol. 2018. PMID: 29669828 Free PMC article.
-
Domain Interaction Studies of Herpes Simplex Virus 1 Tegument Protein UL16 Reveal Its Interaction with Mitochondria.J Virol. 2017 Jan 3;91(2):e01995-16. doi: 10.1128/JVI.01995-16. Print 2017 Jan 15. J Virol. 2017. PMID: 27847362 Free PMC article.
-
A Conserved Leucine Zipper Motif in Gammaherpesvirus ORF52 Is Critical for Distinct Microtubule Rearrangements.J Virol. 2017 Aug 10;91(17):e00304-17. doi: 10.1128/JVI.00304-17. Print 2017 Sep 1. J Virol. 2017. PMID: 28615210 Free PMC article.
References
-
- Adams R, Cunningham C, Davison MD, MacLean CA, Davison AJ. 1998. Characterization of the protein encoded by gene UL49A of herpes simplex virus type 1. J. Gen. Virol. 79:813–823 - PubMed
-
- Balan P, et al. 1994. An analysis of the in vitro and in vivo phenotypes of mutants of herpes simplex virus type 1 lacking glycoproteins gG, gE, gI or the putative gJ. J. Gen. Virol. 75:1245–1258 - PubMed
-
- Chi JH, Harley CA, Mukhopadhyay A, Wilson DW. 2005. The cytoplasmic tail of herpes simplex virus envelope glycoprotein D binds to the tegument protein VP22 and to capsids. J. Gen. Virol. 86:253–261 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

