NS5A sequence heterogeneity of hepatitis C virus genotype 4a predicts clinical outcome of pegylated-interferon-ribavirin therapy in Egyptian patients
- PMID: 22993188
- PMCID: PMC3502985
- DOI: 10.1128/JCM.02109-12
NS5A sequence heterogeneity of hepatitis C virus genotype 4a predicts clinical outcome of pegylated-interferon-ribavirin therapy in Egyptian patients
Abstract
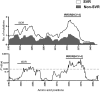
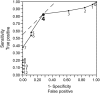
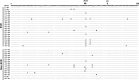
Hepatitis C virus genotype 4 (HCV-4) is the cause of approximately 20% of the 180 million cases of chronic hepatitis C in the world. HCV-4 infection is common in the Middle East and Africa, with an extraordinarily high prevalence in Egypt. Viral genetic polymorphisms, especially within core and NS5A regions, have been implicated in influencing the response to pegylated-interferon and ribavirin (PEG-IFN/RBV) combination therapy in HCV-1 infection. However, this has not been confirmed in HCV-4 infection. Here, we investigated the impact of heterogeneity of NS5A and core proteins of HCV-4, mostly subtype HCV-4a, on the clinical outcomes of 43 Egyptian patients treated with PEG-IFN/RBV. Sliding window analysis over the carboxy terminus of NS5A protein identified the IFN/RBV resistance-determining region (IRRDR) as the most prominent region associated with sustained virological response (SVR). Indeed, 21 (84%) of 25 patients with SVR, but only 5 (28%) of 18 patients with non-SVR, were infected with HCV having IRRDR with 4 or more mutations (IRRDR ≥ 4) (P = 0.0004). Multivariate analysis identified IRRDR ≥ 4 as an independent SVR predictor. The positive predictive value of IRRDR ≥ 4 for SVR was 81% (21/26; P = 0.002), while its negative predictive value for non-SVR was 76% (13/17; P = 0.02). On the other hand, there was no significant correlation between core protein polymorphisms, either at residue 70 or at residue 91, and treatment outcome. In conclusion, the present results demonstrate for the first time that IRRDR ≥ 4, a viral genetic heterogeneity, would be a useful predictive marker for SVR in HCV-4 infection when treated with PEG-IFN/RBV.
Figures

References
-
- Abdel-Aziz F, et al. 2000. Hepatitis C virus (HCV) infection in a community in the Nile Delta: population description and HCV prevalence. Hepatology 32:111–115 - PubMed
-
- Abdel-Hamid M, et al. 2007. Genetic diversity in hepatitis C virus in Egypt and possible association with hepatocellular carcinoma. J. Gen. Virol. 88:1526–1531 - PubMed
-
- Akuta N, et al. 2009. Association of amino acid substitution pattern in core protein of hepatitis C virus genotype 2a high viral load and virological response to interferon-ribavirin combination therapy. Intervirology 52:301–309 - PubMed
-
- Akuta N, et al. 2007. Predictive factors of early and sustained responses to peginterferon plus ribavirin combination therapy in Japanese patients infected with hepatitis C virus genotype 1b: amino acid substitutions in the core region and low-density lipoprotein cholesterol levels. J. Hepatol. 46:403–410 - PubMed
-
- Akuta N, et al. 2007. Prediction of response to pegylated interferon and ribavirin in hepatitis C by polymorphisms in the viral core protein and very early dynamics of viremia. Intervirology 50:361–368 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

