Dual roles of PARP-1 promote cancer growth and progression
- PMID: 22993403
- PMCID: PMC3519969
- DOI: 10.1158/2159-8290.CD-12-0120
Dual roles of PARP-1 promote cancer growth and progression
Abstract
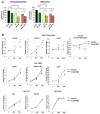
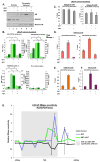
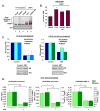
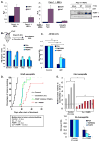
PARP-1 is an abundant nuclear enzyme that modifies substrates by poly(ADP-ribose)-ylation. PARP-1 has well-described functions in DNA damage repair and also functions as a context-specific regulator of transcription factors. With multiple models, data show that PARP-1 elicits protumorigenic effects in androgen receptor (AR)-positive prostate cancer cells, in both the presence and absence of genotoxic insult. Mechanistically, PARP-1 is recruited to sites of AR function, therein promoting AR occupancy and AR function. It was further confirmed in genetically defined systems that PARP-1 supports AR transcriptional function, and that in models of advanced prostate cancer, PARP-1 enzymatic activity is enhanced, further linking PARP-1 to AR activity and disease progression. In vivo analyses show that PARP-1 activity is required for AR function in xenograft tumors, as well as tumor cell growth in vivo and generation and maintenance of castration resistance. Finally, in a novel explant system of primary human tumors, targeting PARP-1 potently suppresses tumor cell proliferation. Collectively, these studies identify novel functions of PARP-1 in promoting disease progression, and ultimately suggest that the dual functions of PARP-1 can be targeted in human prostate cancer to suppress tumor growth and progression to castration resistance.
Significance: These studies introduce a paradigm shift with regard to PARP-1 function in human malignancy, and suggest that the dual functions of PARP-1 in DNA damage repair and transcription factor regulation can be leveraged to suppress pathways critical for promalignant phenotypes in prostate cancer cells by modulation of the DNA damage response and hormone signaling pathways. The combined studies highlight the importance of dual PARP-1 function in malignancy and provide the basis for therapeutic targeting.
©2012 AACR.
Figures

References
-
- Juarez-Salinas H, Sims JL, Jacobson MK. Poly(ADP-ribose) levels in carcinogen-treated cells. Nature. 1979;282:740–741. - PubMed
-
- Benjamin RC, Gill DM. ADP-ribosylation in mammalian cell ghosts. Dependence of poly(ADP-ribose) synthesis on strand breakage in DNA. J Biol Chem. 1980;255:10493–10501. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

