Bacterial Selection during the Formation of Early-Stage Aerobic Granules in Wastewater Treatment Systems Operated Under Wash-Out Dynamics
- PMID: 22993513
- PMCID: PMC3440769
- DOI: 10.3389/fmicb.2012.00332
Bacterial Selection during the Formation of Early-Stage Aerobic Granules in Wastewater Treatment Systems Operated Under Wash-Out Dynamics
Abstract
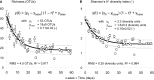
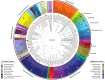
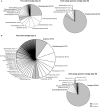
Aerobic granular sludge is attractive for high-rate biological wastewater treatment. Biomass wash-out conditions stimulate the formation of aerobic granules. Deteriorated performances in biomass settling and nutrient removal during start-up have however often been reported. The effect of wash-out dynamics was investigated on bacterial selection, biomass settling behavior, and metabolic activities during the formation of early-stage granules from activated sludge of two wastewater treatment plants (WWTP) over start-up periods of maximum 60 days. Five bubble-column sequencing batch reactors were operated with feast-famine regimes consisting of rapid pulse or slow anaerobic feeding followed by aerobic starvation. Slow-settling fluffy granules were formed when an insufficient superficial air velocity (SAV; 1.8 cm s(-1)) was applied, when the inoculation sludge was taken from a WWTP removing organic matter only, or when reactors were operated at 30°C. Fast-settling dense granules were obtained with 4.0 cm s(-1) SAV, or when the inoculation sludge was taken from a WWTP removing all nutrients biologically. However, only carbon was aerobically removed during start-up. Fluffy granules and dense granules were displaying distinct predominant phylotypes, namely filamentous Burkholderiales affiliates and Zoogloea relatives, respectively. The latter were predominant in dense granules independently from the feeding regime. A combination of insufficient solid retention time and of leakage of acetate into the aeration phase during intensive biomass wash-out was the cause for the proliferation of Zoogloea spp. in dense granules, and for the deterioration of BNR performances. It is however not certain that Zoogloea-like organisms are essential in granule formation. Optimal operation conditions should be elucidated for maintaining a balance between organisms with granulation propensity and nutrient removing organisms in order to form granules with BNR activities in short start-up periods.
Keywords: aerobic granular sludge; bacterial selection; biological wastewater treatment; granule formation; nutrient removal limitations; wash-out dynamics.
Figures




Similar articles
-
Assessment of bacterial and structural dynamics in aerobic granular biofilms.Front Microbiol. 2013 Jul 10;4:175. doi: 10.3389/fmicb.2013.00175. eCollection 2013. Front Microbiol. 2013. PMID: 23847600 Free PMC article.
-
Upflow anaerobic sludge blanket reactor--a review.Indian J Environ Health. 2001 Apr;43(2):1-82. Indian J Environ Health. 2001. PMID: 12397675 Review.
-
Development of aerobic granular sludge under tropical climate conditions: The key role of inoculum adaptation under reduced sludge washout for stable granulation.J Environ Manage. 2019 Jan 15;230:168-182. doi: 10.1016/j.jenvman.2018.09.072. Epub 2018 Oct 3. J Environ Manage. 2019. PMID: 30292012
-
Formation of aerobic granules for the treatment of real and low-strength municipal wastewater using a sequencing batch reactor operated at constant volume.Water Res. 2016 Nov 15;105:341-350. doi: 10.1016/j.watres.2016.09.007. Epub 2016 Sep 9. Water Res. 2016. PMID: 27639343
-
State of the art of aerobic granulation in continuous flow bioreactors.Biotechnol Adv. 2018 Jul-Aug;36(4):1139-1166. doi: 10.1016/j.biotechadv.2018.03.015. Epub 2018 Mar 26. Biotechnol Adv. 2018. PMID: 29597030 Review.
Cited by
-
Identification of protein-degraders in an anaerobic digester by protein stable isotope probing and metagenomics.Appl Microbiol Biotechnol. 2025 Apr 10;109(1):87. doi: 10.1007/s00253-025-13483-5. Appl Microbiol Biotechnol. 2025. PMID: 40204977 Free PMC article.
-
Diversity and abundance of filamentous and non-filamentous "Leptothrix" in global wastewater treatment plants.Appl Environ Microbiol. 2025 Mar 19;91(3):e0148524. doi: 10.1128/aem.01485-24. Epub 2025 Feb 14. Appl Environ Microbiol. 2025. PMID: 39950813 Free PMC article.
-
Multistability and Reversibility of Aerobic Granular Sludge Microbial Communities Upon Changes From Simple to Complex Synthetic Wastewater and Back.Front Microbiol. 2020 Nov 26;11:574361. doi: 10.3389/fmicb.2020.574361. eCollection 2020. Front Microbiol. 2020. PMID: 33324361 Free PMC article.
-
Assessment of bacterial and structural dynamics in aerobic granular biofilms.Front Microbiol. 2013 Jul 10;4:175. doi: 10.3389/fmicb.2013.00175. eCollection 2013. Front Microbiol. 2013. PMID: 23847600 Free PMC article.
-
Changes in bacterial diversity of activated sludge exposed to titanium dioxide nanoparticles.Biodegradation. 2021 Jun;32(3):313-326. doi: 10.1007/s10532-021-09939-w. Epub 2021 Apr 3. Biodegradation. 2021. PMID: 33811584
References
LinkOut - more resources
Full Text Sources
Miscellaneous

