A virological view of innate immune recognition
- PMID: 22994491
- PMCID: PMC3549330
- DOI: 10.1146/annurev-micro-092611-150203
A virological view of innate immune recognition
Abstract
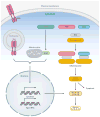
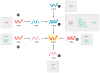
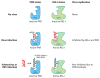
The innate immune system uses multiple strategies to detect viral infections. Because all viruses rely on host cells for their synthesis and propagation, the molecular features used to detect viral infections must be unique to viruses and absent from host cells. Research in the past decade has advanced our understanding of various cell-intrinsic and cell-extrinsic modes of virus recognition. This review examines the innate recognition from the point of view of virus invasion and replication strategies, and places innate sensors in the context of detecting viral genome, replication intermediate, transcriptional by-product, and other viral invasion strategies. On the basis of other unique features common to viral infections, undiscovered areas of virus detection are discussed.
Figures

References
-
- Ablasser A, Bauernfeind F, Hartmann G, Latz E, Fitzgerald KA, Hornung V. RIG-I-dependent sensing of poly(dA:dT) through the induction of an RNA polymerase III-transcribed RNA intermediate. Nat Immunol. 2009;10:1065–72. Shows that Pol III transcripts from dsDNA viruses are recognized by RIG-I (see also Reference 18) - PMC - PubMed
-
- Akira S, Uematsu S, Takeuchi O. Pathogen recognition and innate immunity. Cell. 2006;124:783–801. - PubMed
-
- Alexopoulou L, Holt AC, Medzhitov R, Flavell RA. Recognition of double-stranded RNA and activation of NF-κB by Toll-like receptor 3. Nature. 2001;413:732–38. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

