Prevalence of amino acid mutations in hepatitis C virus core and NS5B regions among Venezuelan viral isolates and comparison with worldwide isolates
- PMID: 22995142
- PMCID: PMC3511240
- DOI: 10.1186/1743-422X-9-214
Prevalence of amino acid mutations in hepatitis C virus core and NS5B regions among Venezuelan viral isolates and comparison with worldwide isolates
Abstract
Background: Recent reports show that R70Q and L/C91M amino acid substitutions in the core from different hepatitis C virus (HCV) genotypes have been associated with variable responses to interferon (IFN) and ribavirin (RBV) therapy, as well to an increase of hepatocellular carcinoma (HCC) risk, liver steatosis and insulin resistance (IR). Mutations in NS5B have also been associated to IFN, RBV, nucleoside and non-nucleoside inhibitors drug resistance. The prevalence of these mutations was studied in HCV RNA samples from chronically HCV-infected drug-naïve patients.
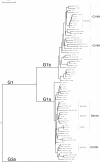
Methods: After amplification of core and NS5B region by nested-PCR, 12 substitutions were analyzed in 266 Venezuelan HCV isolates subtype 1a, 1b, 2a, 2c, 2b, 2j (a subtype frequently found in Venezuela) and 3a (n = 127 and n = 228 for core and NS5B respectively), and compared to isolates from other countries (n = 355 and n = 646 for core and NS5B respectively).
Results: R70Q and L/C91M core substitutions were present exclusively in HCV G1b. Both substitutions were more frequent in American isolates compared to Asian ones (69% versus 26%, p < 0.001 and 75% versus 45%, p < 0.001 respectively). In Venezuelan isolates NS5B D310N substitution was detected mainly in G3a (100%) and G1a (13%), this later with a significantly higher prevalence than in Brazilian isolates (p = 0.03). The NS5B mutations related to IFN/RBV treatment D244N was mainly found in G3a, and Q309R was present in all genotypes, except G2. Resistance to new NS5B inhibitors (C316N) was only detected in 18% of G1b, with a significantly lower prevalence than in Asian isolates, where this polymorphism was surprisingly frequent (p < 0.001).
Conclusions: Genotypical, geographical and regional differences were found in the prevalence of substitutions in HCV core and NS5B proteins. The substitutions found in the Venezuelan G2j type were similar to that found in G2a and G2c isolates. Our results suggest a high prevalence of the R70Q and L/C91M mutations of core protein for G1b and D310N substitution of NS5B protein for the G3a. C316N polymorphism related with resistance to new NS5B inhibitors was only found in G1b. Some of these mutations could be associated with a worse prognosis of the disease in HCV infected patients.
Figures
References
-
- World health organization report June 2011, fact sheet N°164. http://www.who.int/mediacentre/factsheets/fs164/en/
-
- Aguilar MS, Cosson C, Loureiro CL, Devesa M, Martínez J, Villegas L, Flores J, Ludert JE, Noya O, Liprandi F, Pujol FH, Alarcón de Noya B. Prevalence of hepatitis C virus infection in Venezuela assessed by a synthetic peptide-based immunoassay. Ann Trop Med Parasitol. 2001;95:187–195. doi: 10.1080/00034980120042944. - DOI - PubMed
-
- Silva L, Paraná R, Mota E, Cotrim HP, Boënnec-McCurtey ML, Vitvitinsky L, Pádua A, Trepo C, Lyra L. Prevalence of hepatitis C virus in urban and rural populations of northeast brazil - pilot study. Arq Gastroenterol. 1995;32:168–171. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

