Transcription stochasticity of complex gene regulation models
- PMID: 22995487
- PMCID: PMC3446772
- DOI: 10.1016/j.bpj.2012.07.011
Transcription stochasticity of complex gene regulation models
Abstract
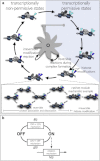
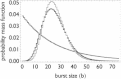
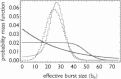
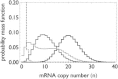
Transcription is regulated by a multitude of factors that concertedly induce genes to switch between activity states. Eukaryotic transcription involves a multitude of complexes that sequentially assemble on chromatin under the influence of transcription factors and the dynamic state of chromatin. Prokaryotic transcription depends on transcription factors, sigma-factors, and, in some cases, on DNA looping. We present a stochastic model of transcription that considers these complex regulatory mechanisms. We coarse-grain the molecular details in such a way that the model can describe a broad class of gene-regulation mechanisms. We solve this model analytically for various measures of stochastic transcription and compare alternative gene-regulation designs. We find that genes with complex multiprotein regulation can have peaked burst-size distributions in contrast to the geometric distributions found for simple models of transcription regulation. Burst-size distributions are, in addition, shaped by mRNA degradation during transcription bursts. We derive the stochastic properties of genes in the limit of deterministic switch times. These genes typically have reduced transcription noise. Severe timescale separation between gene regulation and transcription initiation enhances noise and leads to bimodal mRNA copy number distributions. In general, complex mechanisms for gene regulation lead to nonexponential waiting-time distributions for gene switching and transcription initiation, which typically reduce noise in mRNA copy numbers and burst size. Finally, we discuss that qualitatively different gene regulation models can often fit the same experimental data on single-cell mRNA abundance even though they have qualitatively different burst-size statistics and regulatory parameters.
Copyright © 2012 Biophysical Society. Published by Elsevier Inc. All rights reserved.
Figures


References
-
- Berger S.L. The complex language of chromatin regulation during transcription. Nature. 2007;447:407–412. - PubMed
-
- Weake V.M., Workman J.L. Inducible gene expression: diverse regulatory mechanisms. Nat. Rev. Genet. 2010;11:426–437. - PubMed
-
- Ptashne M., Gann A. Transcriptional activation by recruitment. Nature. 1997;386:569–577. - PubMed
-
- Clapier C.R., Cairns B.R. The biology of chromatin remodeling complexes. Annu. Rev. Biochem. 2009;78:273–304. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

