Robust driving forces for transmembrane helix packing
- PMID: 22995495
- PMCID: PMC3446696
- DOI: 10.1016/j.bpj.2012.08.035
Robust driving forces for transmembrane helix packing
Abstract
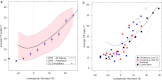
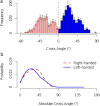
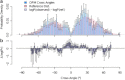
The packing structures of transmembrane helices are traditionally attributed to patterns in residues along the contact surface. In this view, besides keeping the helices confined in the membrane, the bilayer has only a minor effect on the helices structure. Here, we use two different approaches to show that the lipid environment has a crucial effect in determining the cross-angle distribution of packed helices. We analyzed structural data of a membrane proteins database. We show that the distribution of cross angles of helix pairs in this database is statistically indistinguishable from the cross-angle distribution of two noninteracting helices imbedded in the membrane. These results suggest that the cross angle is, to a large extent, determined by the tilt angle of the individual helices. We test this hypothesis using molecular simulations of a coarse-grained model that contains no specific residue interactions. These simulations reproduce the same cross-angle distribution as found in the database. As the tilt angle of a helix is dominated by hydrophobic mismatch between the protein and surrounding lipids, our results indicate that hydrophobic mismatch is the dominant factor guiding the transmembrane helix packing. Other short-range forces might then fine-tune the structure to its final configuration.
Copyright © 2012 Biophysical Society. Published by Elsevier Inc. All rights reserved.
Figures




Similar articles
-
Revisiting hydrophobic mismatch with free energy simulation studies of transmembrane helix tilt and rotation.Biophys J. 2010 Jul 7;99(1):175-83. doi: 10.1016/j.bpj.2010.04.015. Biophys J. 2010. PMID: 20655845 Free PMC article.
-
Analyzing the effects of hydrophobic mismatch on transmembrane α-helices using tryptophan fluorescence spectroscopy.Methods Mol Biol. 2013;1063:95-116. doi: 10.1007/978-1-62703-583-5_5. Methods Mol Biol. 2013. PMID: 23975773
-
Transmembrane helices of membrane proteins may flex to satisfy hydrophobic mismatch.Biochim Biophys Acta. 2007 Mar;1768(3):530-7. doi: 10.1016/j.bbamem.2006.11.018. Epub 2006 Dec 15. Biochim Biophys Acta. 2007. PMID: 17223071
-
Ca2+ -ATPase structure in the E1 and E2 conformations: mechanism, helix-helix and helix-lipid interactions.Biochim Biophys Acta. 2002 Oct 11;1565(2):246-66. doi: 10.1016/s0005-2736(02)00573-4. Biochim Biophys Acta. 2002. PMID: 12409199 Review.
-
Marginally hydrophobic transmembrane α-helices shaping membrane protein folding.Protein Sci. 2015 Jul;24(7):1057-74. doi: 10.1002/pro.2698. Epub 2015 May 30. Protein Sci. 2015. PMID: 25970811 Free PMC article. Review.
Cited by
-
Small scale membrane mechanics.Biomech Model Mechanobiol. 2014 Aug;13(4):697-711. doi: 10.1007/s10237-013-0528-6. Epub 2013 Oct 1. Biomech Model Mechanobiol. 2014. PMID: 24081650 Free PMC article.
-
Structural Symmetry in Membrane Proteins.Annu Rev Biophys. 2015;44:311-37. doi: 10.1146/annurev-biophys-051013-023008. Annu Rev Biophys. 2015. PMID: 26098517 Free PMC article. Review.
-
Conformational Changes in the Epidermal Growth Factor Receptor: Role of the Transmembrane Domain Investigated by Coarse-Grained MetaDynamics Free Energy Calculations.J Am Chem Soc. 2016 Aug 24;138(33):10611-22. doi: 10.1021/jacs.6b05602. Epub 2016 Aug 11. J Am Chem Soc. 2016. PMID: 27459426 Free PMC article.
-
The free energy landscape of dimerization of a membrane protein, NanC.PLoS Comput Biol. 2014 Jan;10(1):e1003417. doi: 10.1371/journal.pcbi.1003417. Epub 2014 Jan 9. PLoS Comput Biol. 2014. PMID: 24415929 Free PMC article.
-
Interactions of amino acid side-chain analogs within membrane environments.J Phys Chem B. 2015 Feb 19;119(7):2877-85. doi: 10.1021/jp511712u. Epub 2015 Feb 6. J Phys Chem B. 2015. PMID: 25621811 Free PMC article.
References
-
- Popot J.L., Engelman D.M. Membrane protein folding and oligomerization: the two-stage model. Biochemistry. 1990;29:4031–4037. - PubMed
-
- White S.H., von Heijne G. The machinery of membrane protein assembly. Curr. Opin. Struct. Biol. 2004;14:397–404. - PubMed
-
- Liang J. Experimental and computational studies of determinants of membrane-protein folding. Curr. Opin. Chem. Biol. 2002;6:878–884. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

