Quantitative and sensitive detection of the SARS-CoV spike protein using bispecific monoclonal antibody-based enzyme-linked immunoassay
- PMID: 22995576
- PMCID: PMC7112864
- DOI: 10.1016/j.jviromet.2012.09.006
Quantitative and sensitive detection of the SARS-CoV spike protein using bispecific monoclonal antibody-based enzyme-linked immunoassay
Abstract
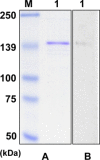
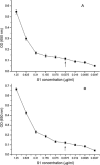
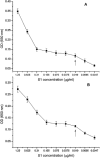
The severe acute respiratory syndrome coronavirus (SARS-CoV) spike protein is known to mediate receptor interaction and immune recognition and thus it is considered as a major target for vaccine design. The spike protein plays an important role in virus entry, virus receptor interactions, and virus tropism. Sensitive diagnosis of SARS is essential for the control of the disease in humans. Recombinant SARS-CoV S1 antigen was produced and purified for the development of monoclonal and bi-specific monoclonal antibodies. The hybridomas secreting anti-S1 antibodies, F26G18 and P136.8D12, were fused respectively with the YP4 hybridoma to generate quadromas. The sandwich ELISA was formed by using F26G18 as a coating antibody and biotinylated F26G18 as a detection antibody with a detection limit of 0.037μg/ml (p<0.02). The same detection limit was found with P136.8D12 as a coating antibody and biotinylated F26G18 as a detection antibody. The sensitivity was improved (detection limit of 0.019μg/ml), however, when using bi-specific monoclonal antibody (F157) as the detection antibody. In conclusion, the method described in this study allows sensitive detection of a recombinant SARS spike protein by sandwich ELISA with bi-specific monoclonal antibody and could be used for the diagnosis of patients suspected with SARS.
Copyright © 2012 Elsevier B.V. All rights reserved.
Figures
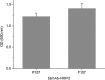
References
-
- Balboni A., Battilani M., Prosperi S. The SARS-like coronaviruses: the role of bats and evolutionary relationships with SARS coronavirus. New Microbiologica. 2012;35:1–16. - PubMed
-
- Colman P.M., Lawrence M.C. The structural biology of type I viral membrane fusion. Nature Reviews Molecular Cell Biology. 2003;4:309–319. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

