The 70S ribosome modulates the ATPase activity of Escherichia coli YchF
- PMID: 22995830
- PMCID: PMC3583859
- DOI: 10.4161/rna.22131
The 70S ribosome modulates the ATPase activity of Escherichia coli YchF
Abstract
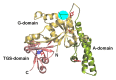
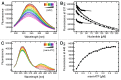
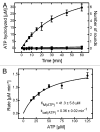
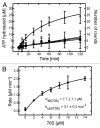
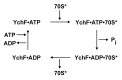
YchF is one of two universally conserved GTPases with unknown cellular function. As a first step toward elucidating YchF's cellular role, we performed a detailed biochemical characterization of the protein from Escherichia coli. Our data from fluorescence titrations not only confirmed the surprising finding that YchFE.coli binds adenine nucleotides more efficiently than guanine nucleotides, but also provides the first evidence suggesting that YchF assumes two distinct conformational states (ATP- and ADP-bound) consistent with the functional cycle of a typical GTPase. Based on an in vivo pull-down experiment using a His-tagged variant of YchF from E. coli (YchFE.coli), we were able to isolate a megadalton complex containing the 70S ribosome. Based on this finding, we report the successful reconstitution of a YchF•70S complex in vitro, revealing an affinity (KD) of the YchFE.coli•ADPNP complex for 70S ribosomes of 3 μM. The in vitro reconstitution data also suggests that the identity of the nucleotide-bound state of YchF (ADP or ATP) modulates its affinity for 70S ribosomes. A detailed Michaelis-Menten analysis of YchF's catalytic activity in the presence and the absence of the 70S ribosome and its subunits revealed for the first time that the 70S ribosome is able to stimulate YchF's ATPase activity (~10-fold), confirming the ribosome as part of the functional cycle of YchF. Our findings taken together with previously reported data for the human homolog of YchF (hOLA1) indicate a high level of evolutionary conservation in the enzymatic properties of YchF and suggest that the ribosome is the main functional partner of YchF not only in bacteria.
Keywords: ATPase activating factor; ATPase activity; Escherichia coli; HAS-ATPase; Michaelis-Menten kinetics; RNA-binding protein; YchF; molecular switch; ribosome; universally conserved P-loop ATPase.
Figures


References
-
- Mittenhuber G. Comparative genomics of prokaryotic GTP-binding proteins (the Era, Obg, EngA, ThdF (TrmE), YchF and YihA families) and their relationship to eukaryotic GTP-binding proteins (the DRG, ARF, RAB, RAN, RAS and RHO families) J Mol Microbiol Biotechnol. 2001;3:21–35. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
