Red fluorescent proteins: chromophore formation and cellular applications
- PMID: 23000031
- PMCID: PMC3737244
- DOI: 10.1016/j.sbi.2012.09.002
Red fluorescent proteins: chromophore formation and cellular applications
Abstract
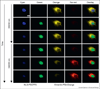
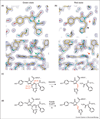
In the last decade, a number of red fluorescent proteins (RFPs) that emit orange, red, and far-red fluorescence have been isolated from anthozoans (corals), and developed through directed molecular evolution. An attractive property possessed by some RFPs is that their red fluorescence can be turned on or modulated by illumination at specific wavelengths. Recent progress in the development of RFPs has been accompanied with detailed studies of chromophore chemistry. A thorough understanding of the molecular mechanisms involved in the post-translational modifications of red chromophores would enable scientists to design RFPs with the desired properties to advance imaging applications. This article provides a broad perspective on the chemistry and applications of RFPs.
Copyright © 2012 Elsevier Ltd. All rights reserved.
Figures



References
-
- Tsien RY. The green fluorescent protein. Annu Rev Biochem. 1998;67:509–544. - PubMed
-
-
Matz MV, Fradkov AF, Labas YA, Savitsky AP, Zaraisky AG, Markelov ML, Lukyanov SA. Fluorescent proteins from nonbioliminescent Anthozoa species. Nat Biotechnol. 1999;17:969–972.A key paper demonstrating that anthozoan animals express spontaneously fluorescent proteins.
-
-
- Chudakov DM, Matz MV, Lukyanov S, Lukyanov KA. Fluorescent proteins and their applications in imaging living cells and tissues. Physiol Rev. 2010;90:1103–1163. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

