The structure of the plk4 cryptic polo box reveals two tandem polo boxes required for centriole duplication
- PMID: 23000383
- PMCID: PMC3496063
- DOI: 10.1016/j.str.2012.08.025
The structure of the plk4 cryptic polo box reveals two tandem polo boxes required for centriole duplication
Abstract
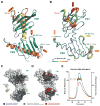
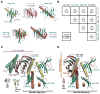
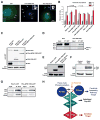
Centrioles are key microtubule polarity determinants. Centriole duplication is tightly controlled to prevent cells from developing multipolar spindles, a situation that promotes chromosomal instability. A conserved component in the duplication pathway is Plk4, a polo kinase family member that localizes to centrioles in M/G1. To limit centriole duplication, Plk4 levels are controlled through trans-autophosphorylation that primes ubiquitination. In contrast to Plks 1-3, Plk4 possesses a unique central region called the "cryptic polo box." Here, we present the crystal structure of this region at 2.3 Å resolution. Surprisingly, the structure reveals two tandem homodimerized polo boxes, PB1-PB2, that form a unique winged architecture. The full PB1-PB2 cassette is required for binding the centriolar protein Asterless as well as robust centriole targeting. Thus, with its C-terminal polo box (PB3), Plk4 has a triple polo box architecture that facilitates oligomerization, targeting, and promotes trans-autophosphorylation, limiting centriole duplication to once per cell cycle.
Copyright © 2012 Elsevier Ltd. All rights reserved.
Figures



Comment in
-
Polo boxes come out of the crypt: a new view of PLK function and evolution.Structure. 2012 Nov 7;20(11):1801-4. doi: 10.1016/j.str.2012.10.008. Structure. 2012. PMID: 23141691
References
-
- Archambault V, Glover DM. Polo-like kinases: conservation and divergence in their functions and regulation. Nat Rev Mol Cell Biol. 2009;10:265–275. - PubMed
-
- Bettencourt-Dias M, Rodrigues-Martins A, Carpenter L, Riparbelli M, Lehmann L, Gatt MK, Carmo N, Balloux F, Callaini G, Glover DM. SAK/PLK4 is required for centriole duplication and flagella development. Curr Biol. 2005;15:2199–2207. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

