Epigenetics of colorectal cancer
- PMID: 23000599
- PMCID: PMC3611241
- DOI: 10.1053/j.gastro.2012.09.032
Epigenetics of colorectal cancer
Abstract
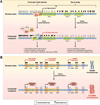
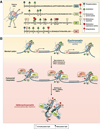
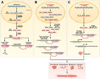
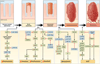
In the early years of the molecular biology revolution, cancer research was mainly focused on genetic changes (ie, those that altered DNA sequences). Although this has been extremely useful as our understanding of the pathogenesis and biology of cancer has grown and matured, there is another realm in tumor development that does not involve changing the sequence of cellular DNA. This field is called "epigenetics" and broadly encompasses changes in the methylation of cytosines in DNA, changes in histone and chromatin structure, and alterations in the expression of microRNAs, which control the stability of many messenger RNAs and serve as "master regulators" of gene expression. This review focuses on the epigenetics of colorectal cancer and illustrates the impact epigenetics has had on this field.
Copyright © 2012 AGA Institute. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Conflicts of interest
The authors disclose no conflicts.
Figures
References
-
- Gardiner-Garden M, Frommer M. CpG islands in vertebrate genomes. J Mol Biol. 1987;196:261–282. - PubMed
-
- Graff JR, Herman JG, Myohanen S, et al. Mapping patterns of CpG island methylation in normal and neoplastic cells implicates both upstream and downstream regions in de novo methylation. J Biol Chem. 1997;272:22322–22329. - PubMed
-
- Ooi SL, Henikoff S. Germline histone dynamics and epigenetics. Curr Opin Cell Biol. 2007;19:257–265. - PubMed
-
- Chadwick BP, Willard HF. Chromatin of the Barr body: histone and non-histone proteins associated with or excluded from the inactive X chromosome. Hum Mol Genet. 2003;12:2167–2178. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

