RELION: implementation of a Bayesian approach to cryo-EM structure determination
- PMID: 23000701
- PMCID: PMC3690530
- DOI: 10.1016/j.jsb.2012.09.006
RELION: implementation of a Bayesian approach to cryo-EM structure determination
Abstract
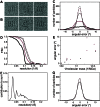
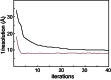
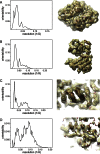
RELION, for REgularized LIkelihood OptimizatioN, is an open-source computer program for the refinement of macromolecular structures by single-particle analysis of electron cryo-microscopy (cryo-EM) data. Whereas alternative approaches often rely on user expertise for the tuning of parameters, RELION uses a Bayesian approach to infer parameters of a statistical model from the data. This paper describes developments that reduce the computational costs of the underlying maximum a posteriori (MAP) algorithm, as well as statistical considerations that yield new insights into the accuracy with which the relative orientations of individual particles may be determined. A so-called gold-standard Fourier shell correlation (FSC) procedure to prevent overfitting is also described. The resulting implementation yields high-quality reconstructions and reliable resolution estimates with minimal user intervention and at acceptable computational costs.
Copyright © 2012 Elsevier Inc. All rights reserved.
Figures

References
-
- Bartolucci C., Lamba D., Grazulis S., Manakova E., Heumann H. Crystal structure of wild-type chaperonin GroEL. Journal of Molecular Biology. 2005;354(4):940–951. - PubMed
-
- Barton B., Rhinow D., Walter A., Schroeder R., Benner G. In-focus electron microscopy of frozen-hydrated biological samples with a Boersch phase plate. Ultramicroscopy. 2011;111(12):1696–1705. - PubMed
-
- Boettcher B., Wynne S.A., Crowther R.A. Determination of the fold of the core protein of hepatitis B virus by electron cryomicroscopy. Nature. 1997;386(6620):88–91. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

