Breast cancer risk-associated SNPs modulate the affinity of chromatin for FOXA1 and alter gene expression
- PMID: 23001124
- PMCID: PMC3483423
- DOI: 10.1038/ng.2416
Breast cancer risk-associated SNPs modulate the affinity of chromatin for FOXA1 and alter gene expression
Abstract
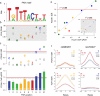
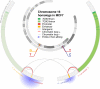
Genome-wide association studies (GWAS) have identified thousands of SNPs that are associated with human traits and diseases. But, because the vast majority of these SNPs are located in non-coding regions of the genome, the mechanisms by which they promote disease risk have remained elusive. Employing a new methodology that combines cistromics, epigenomics and genotype imputation, we annotate the non-coding regions of the genome in breast cancer cells and systematically identify the functional nature of SNPs associated with breast cancer risk. Our results show that breast cancer risk-associated SNPs are enriched in the cistromes of FOXA1 and ESR1 and the epigenome of histone H3 lysine 4 monomethylation (H3K4me1) in a cancer- and cell type-specific manner. Furthermore, the majority of the risk-associated SNPs modulate the affinity of chromatin for FOXA1 at distal regulatory elements, thereby resulting in allele-specific gene expression, which is exemplified by the effect of the rs4784227 SNP on the TOX3 gene within the 16q12.1 risk locus.
Figures



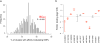

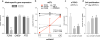
Comment in
-
FOXA1 and breast cancer risk.Nat Genet. 2012 Nov;44(11):1176-7. doi: 10.1038/ng.2449. Nat Genet. 2012. PMID: 23104063
References
-
- Frazer K, Murray S, Schork N, Topol E. Human genetic variation and its contribution to complex traits. Nature Reviews Genetics. 2009;10:241–251. - PubMed
-
- Heintzman N, et al. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nature genetics. 2007;39:311–318. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

