The proto-oncogene MYC is required for selection in the germinal center and cyclic reentry
- PMID: 23001145
- PMCID: PMC3711534
- DOI: 10.1038/ni.2428
The proto-oncogene MYC is required for selection in the germinal center and cyclic reentry
Abstract
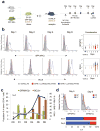
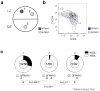
After antigenic challenge, B cells enter the dark zone (DZ) of germinal centers (GCs) to proliferate and hypermutate their immunoglobulin genes. Mutants with greater affinity for the antigen are positively selected in the light zone (LZ) to either differentiate into plasma and memory cells or reenter the DZ. The molecular circuits that govern positive selection in the GC are not known. We show here that the GC reaction required biphasic regulation of expression of the cell-cycle regulator c-Myc that involved its transient induction during early GC commitment, its repression by Bcl-6 in DZ B cells and its reinduction in B cells selected for reentry into the DZ. Inhibition of c-Myc in vivo led to GC collapse, which indicated an essential role for c-Myc in GCs. Our results have implications for the mechanism of GC selection and the role of c-Myc in lymphomagenesis.
Figures





Comment in
-
The case of the missing c-Myc.Nat Immunol. 2012 Nov;13(11):1029-31. doi: 10.1038/ni.2455. Nat Immunol. 2012. PMID: 23080195 No abstract available.
References
-
- MacLennan IC. Germinal centers. Annu Rev Immunol. 1994;12:117–139. - PubMed
-
- Victora GD, Nussenzweig MC. Germinal centers. Annu Rev Immunol. 2012;30:429–457. - PubMed
-
- Allen CD, Okada T, Tang HL, Cyster JG. Imaging of germinal center selection events during affinity maturation. Science. 2007;315:528–531. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

