Removal of a frameshift between the hsdM and hsdS genes of the EcoKI Type IA DNA restriction and modification system produces a new type of system and links the different families of Type I systems
- PMID: 23002145
- PMCID: PMC3510504
- DOI: 10.1093/nar/gks876
Removal of a frameshift between the hsdM and hsdS genes of the EcoKI Type IA DNA restriction and modification system produces a new type of system and links the different families of Type I systems
Abstract
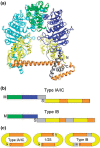
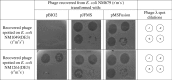
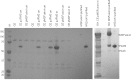
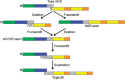
The EcoKI DNA methyltransferase is a trimeric protein comprised of two modification subunits (M) and one sequence specificity subunit (S). This enzyme forms the core of the EcoKI restriction/modification (RM) enzyme. The 3' end of the gene encoding the M subunit overlaps by 1 nt the start of the gene for the S subunit. Translation from the two different open reading frames is translationally coupled. Mutagenesis to remove the frameshift and fuse the two subunits together produces a functional RM enzyme in vivo with the same properties as the natural EcoKI system. The fusion protein can be purified and forms an active restriction enzyme upon addition of restriction subunits and of additional M subunit. The Type I RM systems are grouped into families, IA to IE, defined by complementation, hybridization and sequence similarity. The fusion protein forms an evolutionary intermediate form lying between the Type IA family of RM enzymes and the Type IB family of RM enzymes which have the frameshift located at a different part of the gene sequence.
Figures


Similar articles
-
Fusion of GFP to the M.EcoKI DNA methyltransferase produces a new probe of Type I DNA restriction and modification enzymes.Biochem Biophys Res Commun. 2010 Jul 23;398(2):254-9. doi: 10.1016/j.bbrc.2010.06.069. Epub 2010 Jun 19. Biochem Biophys Res Commun. 2010. PMID: 20599730 Free PMC article.
-
Analysis of type I restriction modification systems in the Neisseriaceae: genetic organization and properties of the gene products.Mol Microbiol. 2001 Sep;41(5):1199-210. doi: 10.1046/j.1365-2958.2001.02587.x. Mol Microbiol. 2001. PMID: 11555298
-
Phosphorylation of Type IA restriction-modification complex enzyme EcoKI on the HsdR subunit.FEMS Microbiol Lett. 2007 May;270(1):171-7. doi: 10.1111/j.1574-6968.2007.00663.x. FEMS Microbiol Lett. 2007. PMID: 17439637
-
Defining domains in type-I restriction and modification enzymes.Gene. 1988 Dec 25;74(1):239-41. doi: 10.1016/0378-1119(88)90295-8. Gene. 1988. PMID: 3074012 Review. No abstract available.
-
The assembly of the EcoKI type I DNA restriction/modification enzyme and its interaction with DNA.Biochem Soc Trans. 1999 Aug;27(4):691-6. doi: 10.1042/bst0270691. Biochem Soc Trans. 1999. PMID: 10917668 Review. No abstract available.
Cited by
-
A model for the evolution of prokaryotic DNA restriction-modification systems based upon the structural malleability of Type I restriction-modification enzymes.Nucleic Acids Res. 2018 Sep 28;46(17):9067-9080. doi: 10.1093/nar/gky760. Nucleic Acids Res. 2018. PMID: 30165537 Free PMC article.
-
CRISPR-Cas3 and type I restriction-modification team up against blaKPC-IncF plasmid transfer in Klebsiella pneumoniae.BMC Microbiol. 2024 Jul 3;24(1):240. doi: 10.1186/s12866-024-03381-7. BMC Microbiol. 2024. PMID: 38961341 Free PMC article.
-
Characterization of the Staphylococcus xylosus methylome reveals a new variant of type I restriction modification system in staphylococci.Front Microbiol. 2023 Mar 8;14:946189. doi: 10.3389/fmicb.2023.946189. eCollection 2023. Front Microbiol. 2023. PMID: 36970683 Free PMC article.
-
Functional C-TERMINALLY ENCODED PEPTIDE (CEP) plant hormone domains evolved de novo in the plant parasite Rotylenchulus reniformis.Mol Plant Pathol. 2016 Oct;17(8):1265-75. doi: 10.1111/mpp.12402. Epub 2016 Jun 6. Mol Plant Pathol. 2016. PMID: 26996971 Free PMC article.
-
Functional comparison of anti-restriction and anti-methylation activities of ArdA, KlcA, and KlcAHS from Klebsiella pneumoniae.Front Cell Infect Microbiol. 2022 Jul 28;12:916547. doi: 10.3389/fcimb.2022.916547. eCollection 2022. Front Cell Infect Microbiol. 2022. PMID: 35967855 Free PMC article.
References
-
- Thomas CM, Nielsen KM. Mechanisms of, and barriers to, horizontal gene transfer between bacteria. Nat. Rev. Microbiol. 2005;3:711–721. - PubMed
-
- Murray NE. Immigration control of DNA in bacteria: self versus non-self. Microbiology. 2002;148:3–20. - PubMed
-
- Tock MR, Dryden DTF. The biology of restriction and anti-restriction. Curr. Op. Microbiol. 2005;8:466–472. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

