Genome-wide activation of latent donor splice sites in stress and disease
- PMID: 23002147
- PMCID: PMC3510495
- DOI: 10.1093/nar/gks834
Genome-wide activation of latent donor splice sites in stress and disease
Abstract
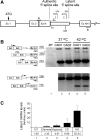
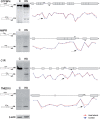
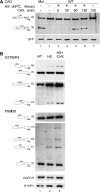
Sequences that conform to the 5' splice site (5'SS) consensus are highly abundant in mammalian introns. Most of these sequences are preceded by at least one in-frame stop codon; thus, their use for splicing would result in pre-maturely terminated aberrant mRNAs. In normally grown cells, such intronic 5'SSs appear not to be selected for splicing. However, under heat shock conditions aberrant splicing involving such latent 5'SSs occurred in a number of specific gene transcripts. Using a splicing-sensitive microarray, we show here that stress-induced (e.g. heat shock) activation of latent splicing is widespread across the human transcriptome, thus highlighting the possibility that latent splicing may underlie certain diseases. Consistent with this notion, our analyses of data from the Gene Expression Omnibus (GEO) revealed widespread activation of latent splicing in cells grown under hypoxia and in certain cancers such as breast cancer and gliomas. These changes were found in thousands of transcripts representing a wide variety of functional groups; among them are genes involved in cell proliferation and differentiation. The GEO analysis also revealed a set of gene transcripts in oligodendroglioma, in which the level of activation of latent splicing increased with the severity of the disease.
Figures