Identification of proteins likely to be involved in morphogenesis, cell division, and signal transduction in Planctomycetes by comparative genomics
- PMID: 23002222
- PMCID: PMC3497475
- DOI: 10.1128/JB.01325-12
Identification of proteins likely to be involved in morphogenesis, cell division, and signal transduction in Planctomycetes by comparative genomics
Abstract
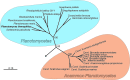
Members of the Planctomycetes clade share many unusual features for bacteria. Their cytoplasm contains membrane-bound compartments, they lack peptidoglycan and FtsZ, they divide by polar budding, and they are capable of endocytosis. Planctomycete genomes have remained enigmatic, generally being quite large (up to 9 Mb), and on average, 55% of their predicted proteins are of unknown function. Importantly, proteins related to the unusual traits of Planctomycetes remain largely unknown. Thus, we embarked on bioinformatic analyses of these genomes in an effort to predict proteins that are likely to be involved in compartmentalization, cell division, and signal transduction. We used three complementary strategies. First, we defined the Planctomycetes core genome and subtracted genes of well-studied model organisms. Second, we analyzed the gene content and synteny of morphogenesis and cell division genes and combined both methods using a "guilt-by-association" approach. Third, we identified signal transduction systems as well as sigma factors. These analyses provide a manageable list of candidate genes for future genetic studies and provide evidence for complex signaling in the Planctomycetes akin to that observed for bacteria with complex life-styles, such as Myxococcus xanthus.
Figures





References
-
- Bauer M, et al. 2004. Archaea-like genes for C1-transfer enzymes in Planctomycetes: phylogenetic implications of their unexpected presence in this phylum. J. Mol. Evol. 59:571–586 - PubMed
-
- Bernander R, Ettema TJ. 2010. FtsZ-less cell division in Archaea and Bacteria. Curr. Opin. Microbiol. 13:747–752 - PubMed
-
- Brochier C, Philippe H. 2002. Phylogeny: a non-hyperthermophilic ancestor for bacteria. Nature 417:244. - PubMed
-
- Butcher BG, Mascher T, Helmann JD. 2008. Environmental sensing and the role of extracytoplasmic function (ECF) sigma factors, p 233–261 In El-Sharoud WM. (ed), Bacterial physiology—a molecular approach Springer-Verlag GmbH, Berlin, Germany
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

