Predicting ion-nucleic acid interactions by energy landscape-guided sampling
- PMID: 23002389
- PMCID: PMC3446742
- DOI: 10.1021/ct300227a
Predicting ion-nucleic acid interactions by energy landscape-guided sampling
Abstract
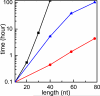
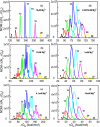
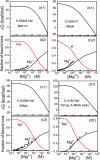
The recently developed Tightly Bound Ion (TBI) model offers improved predictions for ion effect in nucleic acid systems by accounting for ion correlation and fluctuation effects. However, further application of the model to larger systems is limited by the low computational efficiency of the model. Here, we develop a new computational efficient TBI model using free energy landscape-guided sampling method. The method leads to drastic reduction in the computer time by a factor of 50 for RNAs of 50-100 nucleotides long. The improvement in the computational efficiency would be more significant for larger structures. To test the new method, we apply the model to predict the free energies and the number of bound ions for a series of RNA folding systems. The validity of this new model is supported by the nearly exact agreement with the results from the original TBI model and the agreement with the experimental data. The method may pave the way for further applications of the TBI model to treat a broad range of biologically significant systems such as tetraloop-receptor and riboswitches.
Figures
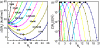
Similar articles
-
Monte Carlo Tightly Bound Ion Model: Predicting Ion-Binding Properties of RNA with Ion Correlations and Fluctuations.J Chem Theory Comput. 2016 Jul 12;12(7):3370-81. doi: 10.1021/acs.jctc.6b00028. Epub 2016 Jun 17. J Chem Theory Comput. 2016. PMID: 27311366 Free PMC article.
-
Exploring the electrostatic energy landscape for tetraloop-receptor docking.Phys Chem Chem Phys. 2014 Apr 14;16(14):6367-75. doi: 10.1039/c3cp53655f. Epub 2013 Dec 10. Phys Chem Chem Phys. 2014. PMID: 24322001 Free PMC article.
-
A New Method to Predict Ion Effects in RNA Folding.Methods Mol Biol. 2017;1632:1-17. doi: 10.1007/978-1-4939-7138-1_1. Methods Mol Biol. 2017. PMID: 28730429 Free PMC article.
-
Theory Meets Experiment: Metal Ion Effects in HCV Genomic RNA Kissing Complex Formation.Front Mol Biosci. 2017 Dec 22;4:92. doi: 10.3389/fmolb.2017.00092. eCollection 2017. Front Mol Biosci. 2017. PMID: 29312955 Free PMC article. Review.
-
An RNA folding motif: GNRA tetraloop-receptor interactions.Q Rev Biophys. 2013 Aug;46(3):223-64. doi: 10.1017/S0033583513000048. Epub 2013 Aug 5. Q Rev Biophys. 2013. PMID: 23915736 Review.
Cited by
-
Reduced model captures Mg(2+)-RNA interaction free energy of riboswitches.Biophys J. 2014 Apr 1;106(7):1508-19. doi: 10.1016/j.bpj.2014.01.042. Biophys J. 2014. PMID: 24703312 Free PMC article.
-
Many-body effect in ion binding to RNA.J Chem Phys. 2014 Aug 7;141(5):055101. doi: 10.1063/1.4890656. J Chem Phys. 2014. PMID: 25106614 Free PMC article.
-
MCTBI: a web server for predicting metal ion effects in RNA structures.RNA. 2017 Aug;23(8):1155-1165. doi: 10.1261/rna.060947.117. Epub 2017 Apr 27. RNA. 2017. PMID: 28450533 Free PMC article.
-
Landscape Zooming toward the Prediction of RNA Cotranscriptional Folding.J Chem Theory Comput. 2022 Mar 8;18(3):2002-2015. doi: 10.1021/acs.jctc.1c01233. Epub 2022 Feb 8. J Chem Theory Comput. 2022. PMID: 35133833 Free PMC article.
-
Predicting Monovalent Ion Correlation Effects in Nucleic Acids.ACS Omega. 2019 Aug 5;4(8):13435-13446. doi: 10.1021/acsomega.9b01689. eCollection 2019 Aug 20. ACS Omega. 2019. PMID: 31460472 Free PMC article.
References
-
- Brion P, Westhof E. Hierarchy and dynamics of RNA folding. Annu. Rev. Biophys. Biomol. Struct. 1997;26:113–137. - PubMed
-
- Tinoco I, Jr, Bustamante C. How RNA folds. J. Mol. Biol. 1999;293:271–281. - PubMed
-
- Sosnick TR, Pan T. RNA folding: models and perspectives. Curr. Opin. Struct. Biol. 2003;13:309–316. - PubMed
-
- Woodson SA. Metal ions and RNA folding: a highly charged topic with a dynamic future. Curr. Opin. Chem. Biol. 2005;9:104–109. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
