The Cbp3-Cbp6 complex coordinates cytochrome b synthesis with bc(1) complex assembly in yeast mitochondria
- PMID: 23007649
- PMCID: PMC3461508
- DOI: 10.1083/jcb.201206040
The Cbp3-Cbp6 complex coordinates cytochrome b synthesis with bc(1) complex assembly in yeast mitochondria
Abstract
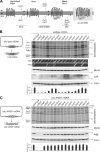
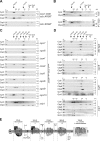
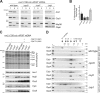
Respiratory chain complexes in mitochondria are assembled from subunits derived from two genetic systems. For example, the bc(1) complex consists of nine nuclear encoded subunits and the mitochondrially encoded subunit cytochrome b. We recently showed that the Cbp3-Cbp6 complex has a dual function for biogenesis of cytochrome b: it is both required for efficient synthesis of cytochrome b and for protection of the newly synthesized protein from proteolysis. Here, we report that Cbp3-Cbp6 also coordinates cytochrome b synthesis with bc(1) complex assembly. We show that newly synthesized cytochrome b assembled through a series of four assembly intermediates. Blocking assembly at early and intermediate steps resulted in sequestration of Cbp3-Cbp6 in a cytochrome b-containing complex, thereby making Cbp3-Cbp6 unavailable for cytochrome b synthesis and thus reducing overall cytochrome b levels. This feedback loop regulates protein synthesis at the inner mitochondrial membrane by directly monitoring the efficiency of bc(1) complex assembly.
Figures




References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

