Proteome variation among Filifactor alocis strains
- PMID: 23008013
- PMCID: PMC4514522
- DOI: 10.1002/pmic.201200211
Proteome variation among Filifactor alocis strains
Abstract
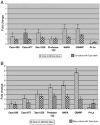
Filifactor alocis, a Gram-positive anaerobic rod, is now considered one of the marker organisms associated with periodontal disease. Although there was heterogeneity in its virulence potential, this bacterium was shown to have virulence properties that may enhance its ability to survive and persist in the periodontal pocket. To gain further insight into a possible mechanism(s) of pathogenesis, the proteome of F. alocis strains was evaluated. Proteins including several proteases, neutrophil-activating protein A and calcium-binding acid repeat protein, were identified in F. alocis. During the invasion of HeLa cells, there was increased expression of several of the genes encoding these proteins in the potentially more virulent F. alocis D-62D compared to F. alocis ATCC 35896, the type strain. A comparative protein in silico analysis of the proteome revealed more cell wall anchoring proteins in the F. alocis D-62D compared to F. alocis ATCC 35896. Their expression was enhanced by coinfection with Porphyromonas gingivalis. Taken together, the variation in the pathogenic potential of the F. alocis strains may be related to the differential expression of several putative virulence factors.
© 2012 WILEY-VCH Verlag GmbH & Co. KGaA, Weinheim.
Conflict of interest statement
The authors have declared no conflict of interest.
Figures




Similar articles
-
Proteome analysis of coinfection of epithelial cells with Filifactor alocis and Porphyromonas gingivalis shows modulation of pathogen and host regulatory pathways.Infect Immun. 2014 Aug;82(8):3261-74. doi: 10.1128/IAI.01727-14. Epub 2014 May 27. Infect Immun. 2014. PMID: 24866790 Free PMC article.
-
Filifactor alocis has virulence attributes that can enhance its persistence under oxidative stress conditions and mediate invasion of epithelial cells by porphyromonas gingivalis.Infect Immun. 2011 Oct;79(10):3872-86. doi: 10.1128/IAI.05631-11. Epub 2011 Aug 8. Infect Immun. 2011. PMID: 21825062 Free PMC article.
-
Role of the Filifactor alocis Hypothetical Protein FA519 in Oxidative Stress Resistance.Microbiol Spectr. 2021 Dec 22;9(3):e0121221. doi: 10.1128/Spectrum.01212-21. Epub 2021 Nov 10. Microbiol Spectr. 2021. PMID: 34756068 Free PMC article.
-
Filifactor alocis: The Newly Discovered Kid on the Block with Special Talents.J Dent Res. 2014 Aug;93(8):725-32. doi: 10.1177/0022034514538283. Epub 2014 Jun 4. J Dent Res. 2014. PMID: 24898946 Free PMC article. Review.
-
Aggregatibacter actinomycetemcomitans and Filifactor alocis: Two exotoxin-producing oral pathogens.Front Oral Health. 2022 Aug 15;3:981343. doi: 10.3389/froh.2022.981343. eCollection 2022. Front Oral Health. 2022. PMID: 36046121 Free PMC article. Review.
Cited by
-
Oral Dysbiotic Communities and Their Implications in Systemic Diseases.Dent J (Basel). 2018 Apr 16;6(2):10. doi: 10.3390/dj6020010. Dent J (Basel). 2018. PMID: 29659479 Free PMC article. Review.
-
FACIN, a Double-Edged Sword of the Emerging Periodontal Pathogen Filifactor alocis: A Metabolic Enzyme Moonlighting as a Complement Inhibitor.J Immunol. 2016 Oct 15;197(8):3245-3259. doi: 10.4049/jimmunol.1600739. Epub 2016 Sep 16. J Immunol. 2016. PMID: 27638863 Free PMC article.
-
Filifactor alocis Promotes Neutrophil Degranulation and Chemotactic Activity.Infect Immun. 2016 Nov 18;84(12):3423-3433. doi: 10.1128/IAI.00496-16. Print 2016 Dec. Infect Immun. 2016. PMID: 27647870 Free PMC article.
-
Proteomic Characterization of the Oral Pathogen Filifactor alocis Reveals Key Inter-Protein Interactions of Its RTX Toxin: FtxA.Pathogens. 2022 May 17;11(5):590. doi: 10.3390/pathogens11050590. Pathogens. 2022. PMID: 35631111 Free PMC article.
-
Proteome analysis of coinfection of epithelial cells with Filifactor alocis and Porphyromonas gingivalis shows modulation of pathogen and host regulatory pathways.Infect Immun. 2014 Aug;82(8):3261-74. doi: 10.1128/IAI.01727-14. Epub 2014 May 27. Infect Immun. 2014. PMID: 24866790 Free PMC article.
References
-
- Saito A, Inagaki S, Kimizuka R, Okuda K, et al. Fusobacterium nucleatum enhances invasion of human gingival epithelial and aortic endothelial cells by Porphyromonas gingivalis. FEMS Immunol Med Microbiol. 2008;54:349–355. - PubMed
-
- Saito A, Inagaki S, Ishihara K. Differential ability of periodontopathic bacteria to modulate invasion of human gingival epithelial cells by Porphyromonas gingivalis. Microb Pathog. 2009;47:329–333. - PubMed
-
- Amano A. Disruption of epithelial barrier and impairment of cellular function by Porphyromonas gingivalis. Front Biosci. 2007;12:3965–3974. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

