Engineering of acetyl-CoA metabolism for the improved production of polyhydroxybutyrate in Saccharomyces cerevisiae
- PMID: 23009357
- PMCID: PMC3519744
- DOI: 10.1186/2191-0855-2-52
Engineering of acetyl-CoA metabolism for the improved production of polyhydroxybutyrate in Saccharomyces cerevisiae
Abstract
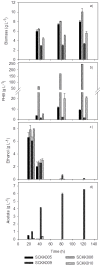
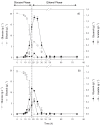
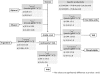
Through metabolic engineering microorganisms can be engineered to produce new products and further produce these with higher yield and productivities. Here, we expressed the bacterial polyhydroxybutyrate (PHB) pathway in the yeast Saccharomyces cerevisiae and we further evaluated the effect of engineering the formation of acetyl coenzyme A (acetyl-CoA), an intermediate of the central carbon metabolism and precursor of the PHB pathway, on heterologous PHB production by yeast. We engineered the acetyl-CoA metabolism by co-transformation of a plasmid containing genes for native S. cerevisiae alcohol dehydrogenase (ADH2), acetaldehyde dehydrogenase (ALD6), acetyl-CoA acetyltransferase (ERG10) and a Salmonella enterica acetyl-CoA synthetase variant (acsL641P), resulting in acetoacetyl-CoA overproduction, together with a plasmid containing the PHB pathway genes coding for acetyl-CoA acetyltransferase (phaA), NADPH-linked acetoacetyl-CoA reductase (phaB) and poly(3-hydroxybutyrate) polymerase (phaC) from Ralstonia eutropha H16. Introduction of the acetyl-CoA plasmid together with the PHB plasmid, improved the productivity of PHB more than 16 times compared to the reference strain used in this study, as well as it reduced the specific product formation of side products.
Figures

Similar articles
-
Improving biobutanol production in engineered Saccharomyces cerevisiae by manipulation of acetyl-CoA metabolism.J Ind Microbiol Biotechnol. 2013 Sep;40(9):1051-6. doi: 10.1007/s10295-013-1296-0. Epub 2013 Jun 13. J Ind Microbiol Biotechnol. 2013. PMID: 23760499
-
Roles of multiple acetoacetyl coenzyme A reductases in polyhydroxybutyrate biosynthesis in Ralstonia eutropha H16.J Bacteriol. 2010 Oct;192(20):5319-28. doi: 10.1128/JB.00207-10. Epub 2010 Aug 20. J Bacteriol. 2010. PMID: 20729355 Free PMC article.
-
Anaerobic poly-3-D-hydroxybutyrate production from xylose in recombinant Saccharomyces cerevisiae using a NADH-dependent acetoacetyl-CoA reductase.Microb Cell Fact. 2016 Nov 18;15(1):197. doi: 10.1186/s12934-016-0598-0. Microb Cell Fact. 2016. PMID: 27863495 Free PMC article.
-
Engineering cytosolic acetyl-coenzyme A supply in Saccharomyces cerevisiae: Pathway stoichiometry, free-energy conservation and redox-cofactor balancing.Metab Eng. 2016 Jul;36:99-115. doi: 10.1016/j.ymben.2016.03.006. Epub 2016 Mar 23. Metab Eng. 2016. PMID: 27016336 Review.
-
Metabolism and strategies for enhanced supply of acetyl-CoA in Saccharomyces cerevisiae.Bioresour Technol. 2021 Dec;342:125978. doi: 10.1016/j.biortech.2021.125978. Epub 2021 Sep 20. Bioresour Technol. 2021. PMID: 34598073 Review.
Cited by
-
Triggering respirofermentative metabolism in the crabtree-negative yeast Pichia guilliermondii by disrupting the CAT8 gene.Appl Environ Microbiol. 2014 Jul;80(13):3879-87. doi: 10.1128/AEM.00854-14. Epub 2014 Apr 18. Appl Environ Microbiol. 2014. PMID: 24747899 Free PMC article.
-
Metabolic engineering of lipid pathways in Saccharomyces cerevisiae and staged bioprocess for enhanced lipid production and cellular physiology.J Ind Microbiol Biotechnol. 2018 Aug;45(8):707-717. doi: 10.1007/s10295-018-2046-0. Epub 2018 May 26. J Ind Microbiol Biotechnol. 2018. PMID: 29804179
-
In Vivo Validation of In Silico Predicted Metabolic Engineering Strategies in Yeast: Disruption of α-Ketoglutarate Dehydrogenase and Expression of ATP-Citrate Lyase for Terpenoid Production.PLoS One. 2015 Dec 23;10(12):e0144981. doi: 10.1371/journal.pone.0144981. eCollection 2015. PLoS One. 2015. PMID: 26701782 Free PMC article.
-
Glucose-based microbial production of the hormone melatonin in yeast Saccharomyces cerevisiae.Biotechnol J. 2016 May;11(5):717-24. doi: 10.1002/biot.201500143. Epub 2016 Jan 25. Biotechnol J. 2016. PMID: 26710256 Free PMC article.
-
Metabolic pathway engineering for fatty acid ethyl ester production in Saccharomyces cerevisiae using stable chromosomal integration.J Ind Microbiol Biotechnol. 2015 Mar;42(3):477-86. doi: 10.1007/s10295-014-1540-2. Epub 2014 Nov 25. J Ind Microbiol Biotechnol. 2015. PMID: 25422103
References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

