Smelt was the likely beneficiary of an antifreeze gene laterally transferred between fishes
- PMID: 23009612
- PMCID: PMC3499448
- DOI: 10.1186/1471-2148-12-190
Smelt was the likely beneficiary of an antifreeze gene laterally transferred between fishes
Abstract
Background: Type II antifreeze protein (AFP) from the rainbow smelt, Osmerus mordax, is a calcium-dependent C-type lectin homolog, similar to the AFPs from herring and sea raven. While C-type lectins are ubiquitous, type II AFPs are only found in a few species in three widely separated branches of teleost fishes. Furthermore, several other non-homologous AFPs are found in intervening species. We have previously postulated that this sporadic distribution has resulted from lateral gene transfer. The alternative hypothesis, that the AFP evolved from a lectin present in a shared ancestor and that this gene was lost in most species, is not favored because both the exon and intron sequences are highly conserved.
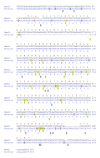
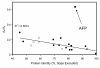
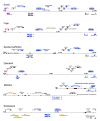
Results: Here we have sequenced and annotated a 160 kb smelt BAC clone containing a centrally-located AFP gene along with 14 other genes. Quantitative PCR indicates that there is but a single copy of this gene within the smelt genome, which is atypical for fish AFP genes. The corresponding syntenic region has been identified and searched in a number of other species and found to be devoid of lectin or AFP sequences. Unlike the introns of the AFP gene, the intronic sequences of the flanking genes are not conserved between species. As well, the rate and pattern of mutation in the AFP gene are radically different from those seen in other smelt and herring genes.
Conclusions: These results provide stand-alone support for an example of lateral gene transfer between vertebrate species. They should further inform the debate about genetically modified organisms by showing that gene transfer between 'higher' eukaryotes can occur naturally. Analysis of the syntenic regions from several fishes strongly suggests that the smelt acquired the AFP gene from the herring.
Figures



Similar articles
-
Lateral transfer of a lectin-like antifreeze protein gene in fishes.PLoS One. 2008 Jul 9;3(7):e2616. doi: 10.1371/journal.pone.0002616. PLoS One. 2008. PMID: 18612417 Free PMC article.
-
Type II antifreeze protein from a mid-latitude freshwater fish, Japanese smelt (Hypomesus nipponensis).Biosci Biotechnol Biochem. 2003 Mar;67(3):461-6. doi: 10.1271/bbb.67.461. Biosci Biotechnol Biochem. 2003. PMID: 12723591
-
Herring antifreeze protein: primary structure and evidence for a C-type lectin evolutionary origin.Mol Mar Biol Biotechnol. 1993 Feb;2(1):20-7. Mol Mar Biol Biotechnol. 1993. PMID: 8364686
-
Control of glycerol production by rainbow smelt (Osmerus mordax) to provide freeze resistance and allow foraging at low winter temperatures.Comp Biochem Physiol B Biochem Mol Biol. 2004 Nov;139(3):347-57. doi: 10.1016/j.cbpc.2004.04.007. Comp Biochem Physiol B Biochem Mol Biol. 2004. PMID: 15544960 Review.
-
Protein evolution revisited.Syst Biol Reprod Med. 2018 Dec;64(6):403-416. doi: 10.1080/19396368.2018.1511764. Epub 2018 Sep 4. Syst Biol Reprod Med. 2018. PMID: 30176752 Review.
Cited by
-
Closing the Gap: Horizontal Transfer of Mariner Transposons between Rhus Gall Aphids and Other Insects.Biology (Basel). 2022 May 10;11(5):731. doi: 10.3390/biology11050731. Biology (Basel). 2022. PMID: 35625459 Free PMC article.
-
Horizontal gene transfer: building the web of life.Nat Rev Genet. 2015 Aug;16(8):472-82. doi: 10.1038/nrg3962. Nat Rev Genet. 2015. PMID: 26184597 Review.
-
Expression of temperature-sensitive ion channel TRPM8 in sperm cells correlates with vertebrate evolution.PeerJ. 2015 Oct 13;3:e1310. doi: 10.7717/peerj.1310. eCollection 2015. PeerJ. 2015. PMID: 26500819 Free PMC article.
-
Sampling the mobile gene pool: innovation via horizontal gene transfer in bacteria.Philos Trans R Soc Lond B Biol Sci. 2017 Dec 5;372(1735):20160424. doi: 10.1098/rstb.2016.0424. Philos Trans R Soc Lond B Biol Sci. 2017. PMID: 29061896 Free PMC article. Review.
-
Rainbow smelt: the unusual case of cryoprotection by sustained glycerol production in an aquatic animal.J Comp Physiol B. 2015 Jul;185(5):487-99. doi: 10.1007/s00360-015-0903-y. Epub 2015 Apr 29. J Comp Physiol B. 2015. PMID: 25921795
References
-
- Duman JG, Olsen TM. Thermal hysteresis protein activity in bacteria, fungi and phylogenetically diverse plants. Cryobiology. 1993;30:322–328. doi: 10.1006/cryo.1993.1031. - DOI
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

