A disintegrin-like and metalloprotease domain containing thrombospondin type 1 motif-like 5 (ADAMTSL5) is a novel fibrillin-1-, fibrillin-2-, and heparin-binding member of the ADAMTS superfamily containing a netrin-like module
- PMID: 23010571
- PMCID: PMC3546522
- DOI: 10.1016/j.matbio.2012.09.003
A disintegrin-like and metalloprotease domain containing thrombospondin type 1 motif-like 5 (ADAMTSL5) is a novel fibrillin-1-, fibrillin-2-, and heparin-binding member of the ADAMTS superfamily containing a netrin-like module
Abstract
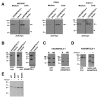
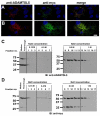
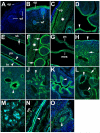
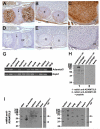
ADAMTS-like proteins are related to ADAMTS metalloproteases by their similarity to ADAMTS ancillary domains. Here, we have characterized ADAMTSL5, a novel member of the superfamily with a unique modular organization that includes a single C-terminal netrin-like (NTR) module. Alternative splicing of ADAMTSL5 at its 5' end generates two transcripts that encode different signal peptides, but the same mature protein. These transcripts differ in their translational efficiency. Recombinant ADAMTSL5 is a secreted, N-glycosylated 60kDa glycoprotein located in the subcellular matrix, on the cell-surface, and in the medium of transfected cells. RT-PCR and western blot analysis of adult mouse tissues showed broad expression. Western blot analysis suggested proteolytic release of the NTR module in transfected cells as well as in some mouse tissues. Immunostaining during mouse organogenesis identified ADAMTSL5 in musculoskeletal tissues such as skeletal muscle, cartilage and bone, as well as in many epithelia. Affinity-chromatography demonstrated heparin-binding of ADAMTSL5 through its NTR-module. Recombinant ADAMTSL5 bound to both fibrillin-1 and fibrillin-2, and co-localized with fibrillin microfibrils in the extracellular matrix of cultured fibroblasts, but without discernible effect on microfibril assembly. ADAMTSL5 is the first family member shown to bind both fibrillin-1 and fibrillin-2. Like other ADAMTS proteins implicated in microfibril biology through identification of human and animal mutations, ADAMTSL5 could have a role in modulating microfibril functions.
Copyright © 2012 International Society of Matrix Biology. Published by Elsevier B.V. All rights reserved.
Figures



References
-
- Bader HL, Ruhe AL, Wang LW, Wong AK, Walsh KF, Packer RA, Mitelman J, Robertson KR, O’Brien DP, Broman KW, Shelton GD, Apte SS, Neff MW. An ADAMTSL2 founder mutation causes Musladin-Lueke Syndrome, a heritable disorder of beagle dogs, featuring stiff skin and joint contractures. PLoS One. 2010;5 - PMC - PubMed
-
- Bekhouche M, Kronenberg D, Vadon-Le Goff S, Bijakowski C, Lim NH, Font B, Kessler E, Colige A, Nagase H, Murphy G, Hulmes DJ, Moali C. Role of the netrin-like domain of procollagen C-proteinase enhancer-1 in the control of metalloproteinase activity. J Biol Chem. 2010;285:15950–15959. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

