Direct visualization of cell division using high-resolution imaging of M-phase of the cell cycle
- PMID: 23011130
- PMCID: PMC3658003
- DOI: 10.1038/ncomms2089
Direct visualization of cell division using high-resolution imaging of M-phase of the cell cycle
Abstract
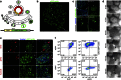
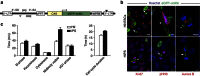
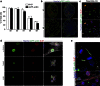
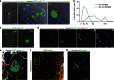
Current approaches to monitor and quantify cell division in live cells, and reliably distinguish between acytokinesis and endoreduplication, are limited and complicate determination of stem cell pool identities. Here we overcome these limitations by generating an in vivo reporter system using the scaffolding protein anillin fused to enhanced green fluorescent protein, to provide high spatiotemporal resolution of mitotic phase. This approach visualizes cytokinesis and midbody formation as hallmarks of expansion of stem and somatic cells, and enables distinction from cell cycle variations. High-resolution microscopy in embryonic heart and brain tissues of enhanced green fluorescent protein-anillin transgenic mice allows live monitoring of cell division and quantitation of cell cycle kinetics. Analysis of cell division in hearts post injury shows that border zone cardiomyocytes in the infarct respond with increasing ploidy, but not cell division. Thus, the enhanced green fluorescent protein-anillin system enables monitoring and measurement of cell division in vivo and markedly simplifies in vitro analysis in fixed cells.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

