Hydra meiosis reveals unexpected conservation of structural synaptonemal complex proteins across metazoans
- PMID: 23012415
- PMCID: PMC3478637
- DOI: 10.1073/pnas.1206875109
Hydra meiosis reveals unexpected conservation of structural synaptonemal complex proteins across metazoans
Abstract
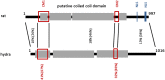
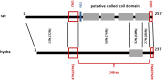
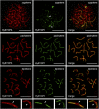
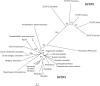
The synaptonemal complex (SC) is a key structure of meiosis, mediating the stable pairing (synapsis) of homologous chromosomes during prophase I. Its remarkable tripartite structure is evolutionarily well conserved and can be found in almost all sexually reproducing organisms. However, comparison of the different SC protein components in the common meiosis model organisms Saccharomyces cerevisiae, Arabidopsis thaliana, Caenorhabditis elegans, Drosophila melanogaster, and Mus musculus revealed no sequence homology. This discrepancy challenged the hypothesis that the SC arose only once in evolution. To pursue this matter we focused on the evolution of SYCP1 and SYCP3, the two major structural SC proteins of mammals. Remarkably, our comparative bioinformatic and expression studies revealed that SYCP1 and SYCP3 are also components of the SC in the basal metazoan Hydra. In contrast to previous assumptions, we therefore conclude that SYCP1 and SYCP3 form monophyletic groups of orthologous proteins across metazoans.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Villeneuve AM, Hillers KJ. Whence meiosis? Cell. 2001;106:647–650. - PubMed
-
- Hunter N. Synaptonemal complexities and commonalities. Mol Cell. 2003;12:533–535. - PubMed
-
- Handel MA, Schimenti JC. Genetics of mammalian meiosis: Regulation, dynamics and impact on fertility. Nat Rev Genet. 2010;11:124–136. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

