Structure of the cleavage-activated prefusion form of the parainfluenza virus 5 fusion protein
- PMID: 23012473
- PMCID: PMC3478641
- DOI: 10.1073/pnas.1213802109
Structure of the cleavage-activated prefusion form of the parainfluenza virus 5 fusion protein
Abstract
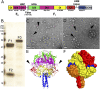
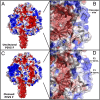
The paramyxovirus parainfluenza virus 5 (PIV5) enters cells by fusion of the viral envelope with the plasma membrane through the concerted action of the fusion (F) protein and the receptor binding protein hemagglutinin-neuraminidase. The F protein folds initially to form a trimeric metastable prefusion form that is triggered to undergo large-scale irreversible conformational changes to form the trimeric postfusion conformation. It is thought that F refolding couples the energy released with membrane fusion. The F protein is synthesized as a precursor (F0) that must be cleaved by a host protease to form a biologically active molecule, F1,F2. Cleavage of F protein is a prerequisite for fusion and virus infectivity. Cleavage creates a new N terminus on F1 that contains a hydrophobic region, known as the FP, which intercalates target membranes during F protein refolding. The crystal structure of the soluble ectodomain of the uncleaved form of PIV5 F is known; here we report the crystal structure of the cleavage-activated prefusion form of PIV5 F. The structure shows minimal movement of the residues adjacent to the protease cleavage site. Most of the hydrophobic FP residues are buried in the uncleaved F protein, and only F103 at the newly created N terminus becomes more solvent-accessible after cleavage. The conformational freedom of the charged arginine residues that compose the protease recognition site increases on cleavage of F protein.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



Comment in
-
Structure of the primed paramyxovirus fusion protein.Proc Natl Acad Sci U S A. 2012 Oct 9;109(41):16404-5. doi: 10.1073/pnas.1214903109. Epub 2012 Oct 1. Proc Natl Acad Sci U S A. 2012. PMID: 23027936 Free PMC article. No abstract available.
References
-
- Lamb RA, Parks GD. Paramyxoviridae: The viruses and their replication. In: Knipe DM, Howley PM, editors. Fields Virology. 5th Ed. Philadelphia: Lippincott Williams & Wilkins; 2007. pp. 1449–1496.
-
- Jardetzky TS, Lamb RA. Virology: A class act. Nature. 2004;427:307–308. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

