Guiding the design of synthetic DNA-binding molecules with massively parallel sequencing
- PMID: 23013524
- PMCID: PMC3483022
- DOI: 10.1021/ja308888c
Guiding the design of synthetic DNA-binding molecules with massively parallel sequencing
Abstract
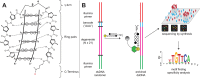
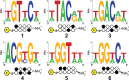
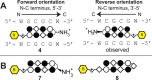
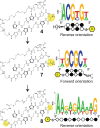
Genomic applications of DNA-binding molecules require an unbiased knowledge of their high affinity sites. We report the high-throughput analysis of pyrrole-imidazole polyamide DNA-binding specificity in a 10(12)-member DNA sequence library using affinity purification coupled with massively parallel sequencing. We find that even within this broad context, the canonical pairing rules are remarkably predictive of polyamide DNA-binding specificity. However, this approach also allows identification of unanticipated high affinity DNA-binding sites in the reverse orientation for polyamides containing β/Im pairs. These insights allow the redesign of hairpin polyamides with different turn units capable of distinguishing 5'-WCGCGW-3' from 5'-WGCGCW-3'. Overall, this study displays the power of high-throughput methods to aid the optimal targeting of sequence-specific minor groove binding molecules, an essential underpinning for biological and nanotechnological applications.
Figures
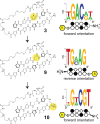
References
-
- Dervan P. B.; Edelson B. S. Curr. Opin. Struct. Biol. 2003, 13, 284–99. - PubMed
-
- Wade W. S.; Mrksich M. L.; Dervan P. B. J. Am. Chem. Soc. 1992, 114, 8783–94.
-
- White S.; Szewczyk J. W.; Turner J. M.; Baird E. E.; Dervan P. B. Nature 1998, 391, 468–71. - PubMed
-
- White S.; Baird E. E.; Dervan P. B. J. Am. Chem. Soc. 1997, 119, 8756–65.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

