Decreased NK cells in patients with head and neck cancer determined in archival DNA
- PMID: 23014525
- PMCID: PMC3500449
- DOI: 10.1158/1078-0432.CCR-12-1008
Decreased NK cells in patients with head and neck cancer determined in archival DNA
Abstract
Purpose: Natural killer (NK) cells are a key element of the innate immune system implicated in human cancer. To examine NK cell levels in archived bloods from a study of human head and neck squamous cell carcinoma (HNSCC), a new DNA-based quantification method was developed.
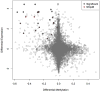
Experimental design: NK cell-specific DNA methylation was identified by analyzing DNA methylation and mRNA array data from purified blood leukocyte subtypes (NK, T, B, monocytes, granulocytes), and confirmed via pyrosequencing and quantitative methylation specific PCR (qMSP). NK cell levels in archived whole blood DNA from 122 HNSCC patients and 122 controls were assessed by qMSP.
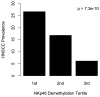
Results: Pyrosequencing and qMSP confirmed that a demethylated DNA region in NKp46 distinguishes NK cells from other leukocytes, and serves as a quantitative NK cell marker. Demethylation of NKp46 was significantly lower in HNSCC patient bloods compared with controls (P < 0.001). Individuals in the lowest NK tertile had over 5-fold risk of being a HNSCC case, controlling for age, gender, HPV16 status, cigarette smoking, alcohol consumption, and BMI (OR = 5.6, 95% CI, 2.0 to 17.4). Cases did not show differences in NKp46 demethylation based on tumor site or stage.
Conclusions: The results of this study indicate a significant depression in NK cells in HNSCC patients that is unrelated to exposures associated with the disease. DNA methylation biomarkers of NK cells represent an alternative to conventional flow cytometry that can be applied in a wide variety of clinical and epidemiologic settings including archival blood specimens.
©2012 AACR.
Conflict of interest statement
Figures


References
-
- Natoli G. Maintaining cell identity through global control of genomic organization. Immunity. 33:12–24. - PubMed
-
- Wieczorek G, Asemissen A, Model F, Turbachova I, Floess S, Liebenberg V, et al. Quantitative DNA methylation analysis of FOXP3 as a new method for counting regulatory T cells in peripheral blood and solid tissue. Cancer Res. 2009;69:599–608. - PubMed
-
- Khavari DA, Sen GL, Rinn JL. DNA methylation and epigenetic control of cellular differentiation. Cell Cycle. 2010;9:3880–3. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

