Identification of and screening for human Helicobacter cinaedi infections and carriers via nested PCR
- PMID: 23015666
- PMCID: PMC3502999
- DOI: 10.1128/JCM.01622-12
Identification of and screening for human Helicobacter cinaedi infections and carriers via nested PCR
Abstract
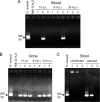
Helicobacter cinaedi is the most frequently reported enterohepatic Helicobacter species isolated from humans. Earlier research suggested that certain patients with H. cinaedi infection may remain undiagnosed or incorrectly diagnosed because of difficulties in detecting the bacteria by conventional culture methods. Here, we report a nested PCR assay that rapidly detects the cytolethal distending toxin gene (cdt) of H. cinaedi with high specificity and sensitivity. Specificity of the assay was validated by using different species of Helicobacter and Campylobacter, as well as known H. cinaedi-positive and -negative samples. The sensitivity of detection for the cdt gene in the assay was 10(2) CFU/ml urine or 10(2) CFU/10(5) infected RAW 264.7 cells. In an H. cinaedi-infected mouse model, the cdt gene of H. cinaedi was effectively detected via the assay with urine (6/7), stool (2/3), and blood (2/6) samples. Importantly, it detected H. cinaedi in blood, urine, and stool samples from one patient with a suspected H. cinaedi infection and three patients with known infections. The assay was further used clinically to follow up two H. cinaedi-infected patients after antibiotic treatment. Stool samples from these two patients evaluated by nested PCR after antibiotic therapy showed clearance of bacterial DNA. Finally, analysis of stool specimens from healthy volunteers showed occasional positive reactions (4/30) to H. cinaedi DNA, which suggests intestinal colonization by H. cinaedi in healthy subjects. In conclusion, this nested PCR assay may be useful for the rapid diagnosis, antimicrobial treatment evaluation, and epidemiological study of H. cinaedi infection.
Figures



References
-
- Fennell CL, et al. 1984. Characterization of Campylobacter-like organisms isolated from homosexual men. J. Infect. Dis. 149:58–66 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

