Differential activation of cellular DNA damage responses by replication-defective and replication-competent adenovirus mutants
- PMID: 23015708
- PMCID: PMC3503084
- DOI: 10.1128/JVI.01757-12
Differential activation of cellular DNA damage responses by replication-defective and replication-competent adenovirus mutants
Abstract
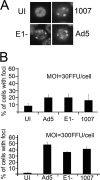
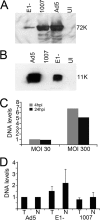
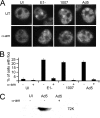
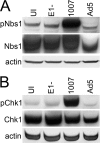
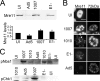
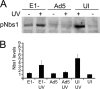
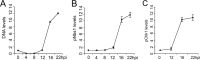
Adenovirus (Ad) mutants that lack early region 4 (E4) activate the phosphorylation of cellular DNA damage response proteins. In wild-type Ad type 5 (Ad5) infections, E1b and E4 proteins target the cellular DNA repair protein Mre11 for redistribution and degradation, thereby interfering with its ability to activate phosphorylation cascades important during DNA repair. The characteristics of Ad infection that activate cellular DNA repair processes are not yet well understood. We investigated the activation of DNA damage responses by a replication-defective Ad vector (AdRSVβgal) that lacks E1 and fails to produce the immediate-early E1a protein. E1a is important for activating early gene expression from the other viral early transcription units, including E4. AdRSVβgal can deliver its genome to the cell, but it is subsequently deficient for viral early gene expression and DNA replication. We studied the ability of AdRSVβgal-infected cells to induce cellular DNA damage responses. AdRSVβgal infection does activate formation of foci containing the Mdc1 protein. However, AdRSVβgal fails to activate phosphorylation of the damage response proteins Nbs1 and Chk1. We found that viral DNA replication is important for Nbs1 phosphorylation, suggesting that this step in the viral life cycle may provide an important trigger for activating at least some DNA repair proteins.
Figures

Similar articles
-
Nbs1-dependent binding of Mre11 to adenovirus E4 mutant viral DNA is important for inhibiting DNA replication.Virology. 2008 Apr 25;374(1):11-22. doi: 10.1016/j.virol.2007.12.034. Epub 2008 Jan 29. Virology. 2008. PMID: 18234271 Free PMC article.
-
DNA-PK phosphorylation at Ser2056 during adenovirus E4 mutant infection is promoted by viral DNA replication and independent of the MRN complex.Virology. 2022 Jan 2;565:82-95. doi: 10.1016/j.virol.2021.10.011. Epub 2021 Nov 2. Virology. 2022. PMID: 34768112
-
Temporal regulation of the Mre11-Rad50-Nbs1 complex during adenovirus infection.J Virol. 2009 May;83(9):4565-73. doi: 10.1128/JVI.00042-09. Epub 2009 Feb 25. J Virol. 2009. PMID: 19244322 Free PMC article.
-
Adenoviral strategies to overcome innate cellular responses to infection.FEBS Lett. 2019 Dec;593(24):3484-3495. doi: 10.1002/1873-3468.13680. Epub 2019 Nov 26. FEBS Lett. 2019. PMID: 31721176 Free PMC article. Review.
-
Complementation for replication by unrelated animal viruses containing DNA genomes.Microbiol Rev. 1987 Dec;51(4):431-8. doi: 10.1128/mr.51.4.431-438.1987. Microbiol Rev. 1987. PMID: 2830477 Free PMC article. Review. No abstract available.
Cited by
-
Localization of the kinase Ataxia Telangiectasia Mutated to Adenovirus E4 mutant DNA replication centers is important for its inhibitory effect on viral DNA accumulation.Virology. 2019 Jan 15;527:47-56. doi: 10.1016/j.virol.2018.11.003. Epub 2018 Nov 16. Virology. 2019. PMID: 30453211 Free PMC article.
-
En Guard! The Interactions between Adenoviruses and the DNA Damage Response.Viruses. 2020 Sep 7;12(9):996. doi: 10.3390/v12090996. Viruses. 2020. PMID: 32906746 Free PMC article. Review.
-
The kinase activity of ataxia-telangiectasia mutated interferes with adenovirus E4 mutant DNA replication.J Virol. 2013 Aug;87(15):8687-96. doi: 10.1128/JVI.00376-13. Epub 2013 Jun 5. J Virol. 2013. PMID: 23740981 Free PMC article.
References
-
- Assenmacher N, Hopfner KP. 2004. MRE11/RAD50/NBS1: complex activities. Chromosoma 113:157–166 - PubMed
-
- Bakkenist CJ, Kastan MB. 2003. DNA damage activates ATM through intermolecular autophosphorylation and dimer dissociation. Nature 421:499–506 - PubMed
-
- Berk AJ, Lee F, Harrison T, Williams J, Sharp PA. 1979. Pre-early adenovirus 5 gene product regulates synthesis of early viral messenger RNAs. Cell 17:935–944 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

