Propionibacterium acnes bacteriophages display limited genetic diversity and broad killing activity against bacterial skin isolates
- PMID: 23015740
- PMCID: PMC3448167
- DOI: 10.1128/mBio.00279-12
Propionibacterium acnes bacteriophages display limited genetic diversity and broad killing activity against bacterial skin isolates
Abstract
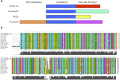
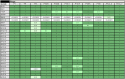
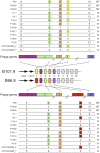
Investigation of the human microbiome has revealed diverse and complex microbial communities at distinct anatomic sites. The microbiome of the human sebaceous follicle provides a tractable model in which to study its dominant bacterial inhabitant, Propionibacterium acnes, which is thought to contribute to the pathogenesis of the human disease acne. To explore the diversity of the bacteriophages that infect P. acnes, 11 P. acnes phages were isolated from the sebaceous follicles of donors with healthy skin or acne and their genomes were sequenced. Comparative genomic analysis of the P. acnes phage population, which spans a 30-year temporal period and a broad geographic range, reveals striking similarity in terms of genome length, percent GC content, nucleotide identity (>85%), and gene content. This was unexpected, given the far-ranging diversity observed in virtually all other phage populations. Although the P. acnes phages display a broad host range against clinical isolates of P. acnes, two bacterial isolates were resistant to many of these phages. Moreover, the patterns of phage resistance correlate closely with the presence of clustered regularly interspaced short palindromic repeat elements in the bacteria that target a specific subset of phages, conferring a system of prokaryotic innate immunity. The limited diversity of the P. acnes bacteriophages, which may relate to the unique evolutionary constraints imposed by the lipid-rich anaerobic environment in which their bacterial hosts reside, points to the potential utility of phage-based antimicrobial therapy for acne.
Importance: Propionibacterium acnes is a dominant member of the skin microflora and has also been implicated in the pathogenesis of acne; however, little is known about the bacteriophages that coexist with and infect this bacterium. Here we present the novel genome sequences of 11 P. acnes phages, thereby substantially increasing the amount of available genomic information about this phage population. Surprisingly, we find that, unlike other well-studied bacteriophages, P. acnes phages are highly homogeneous and show a striking lack of genetic diversity, which is perhaps related to their unique and restricted habitat. They also share a broad ability to kill clinical isolates of P. acnes; phage resistance is not prevalent, but when detected, it appears to be conferred by chromosomally encoded immunity elements within the host genome. We believe that these phages display numerous features that would make them ideal candidates for the development of a phage-based therapy for acne.
Figures

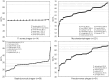

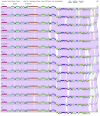
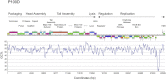
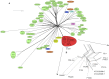
References
-
- Hatfull GF, Science Education Alliance Phage Hunters Advancing Genomics and Evolutionary Science Program, KwaZulu-Natal Research Institute for Tuberculosis and HIV Mycobacterial Genetics Course Students, Phage Hunters Integrating Research and Education Program. 2012. Complete genome sequences of 138 mycobacteriophages. J. Virol. 86:2382–2384 - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

