Evolution of the Arabidopsis telomerase RNA
- PMID: 23015808
- PMCID: PMC3449308
- DOI: 10.3389/fgene.2012.00188
Evolution of the Arabidopsis telomerase RNA
Abstract
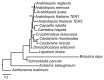
The telomerase reverse transcriptase promotes genome integrity by continually synthesizing a short telomere repeat sequence on chromosome ends. Telomerase is a ribonucleoprotein complex whose integral RNA subunit TER contains a template domain with a sequence complementary to the telomere repeat that is reiteratively copied by the catalytic subunit. Although TER harbors well-conserved secondary structure elements, its nucleotide sequence is highly divergent, even among closely related organisms. Thus, it has been extremely challenging to identify TER orthologs by bioinformatics strategies. Recently, TER was reported in the flowering plant, Arabidopsis thaliana. In contrast to other model organisms, A. thaliana encodes two TER subunits, only one of which is required to maintain telomere tracts in vivo. Here we investigate the evolution of the loci that encode TER in Arabidopsis by comparison to the same locus in its close relatives. We employ a combination of PCR and bioinformatics approaches to identify putative TER loci based on syntenic regions flanking the TER1 and TER2 loci of A. thaliana. Unexpectedly, we discovered that the genomic regions encoding the two A. thaliana TERs occur as a single locus in other Brassicaceae. Moreover, we find striking sequence divergence within the telomere template domain of putative TERs from Brassicaceae, including some orthologous loci that completely lack a template domain. Finally, evolution of the locus is characterized by lineage-specific events rather than changes shared among closely related species. We conclude that the Arabidopsis TER duplication occurred very recently, and further that changes at this locus in other Brassicaceae indicate the process of TER evolution may be different in plants compared with vertebrates and yeast.
Keywords: Brassicaceae; TER; gene duplication; phylogenetics; telomerase.
Figures


References
-
- Autexier C., Lue N. (2006). The structure and function of telomerase reverse transcriptase. Annu. Rev. Biochem. 75 493–517 - PubMed
-
- Beilstein M. A., Al-Shehbaz I. A., Kellogg E. A. (2006). Brassicaceae phylogeny and trichome evolution. Am. J. Bot. 93 607–619 - PubMed
-
- Beilstein M. A., Al-Shehbaz I. A., Mathews S., Kellogg E. A. (2008). Brassicaceae phylogeny inferred from phytochrome A and NDHF sequence data: tribes and trichomes revisited. Am. J. Bot. 95 1307–1327 - PubMed
-
- Chen J. L., Blasco M. A., Greider C. W. (2000). Secondary structure of vertebrate telomerase RNA. Cell 100 503–514 - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

