Integration of ChIP-seq and machine learning reveals enhancers and a predictive regulatory sequence vocabulary in melanocytes
- PMID: 23019145
- PMCID: PMC3483558
- DOI: 10.1101/gr.139360.112
Integration of ChIP-seq and machine learning reveals enhancers and a predictive regulatory sequence vocabulary in melanocytes
Abstract
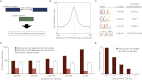
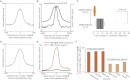
We take a comprehensive approach to the study of regulatory control of gene expression in melanocytes that proceeds from large-scale enhancer discovery facilitated by ChIP-seq; to rigorous validation in silico, in vitro, and in vivo; and finally to the use of machine learning to elucidate a regulatory vocabulary with genome-wide predictive power. We identify 2489 putative melanocyte enhancer loci in the mouse genome by ChIP-seq for EP300 and H3K4me1. We demonstrate that these putative enhancers are evolutionarily constrained, enriched for sequence motifs predicted to bind key melanocyte transcription factors, located near genes relevant to melanocyte biology, and capable of driving reporter gene expression in melanocytes in culture (86%; 43/50) and in transgenic zebrafish (70%; 7/10). Next, using the sequences of these putative enhancers as a training set for a supervised machine learning algorithm, we develop a vocabulary of 6-mers predictive of melanocyte enhancer function. Lastly, we demonstrate that this vocabulary has genome-wide predictive power in both the mouse and human genomes. This study provides deep insight into the regulation of gene expression in melanocytes and demonstrates a powerful approach to the investigation of regulatory sequences that can be applied to other cell types.
Figures





References
-
- Antonellis A, Bennett WR, Menheniott TR, Prasad AB, Lee-Lin SQ, Green ED, Paisley D, Kelsh RN, Pavan WJ, Ward A 2006. Deletion of long-range sequences at Sox10 compromises developmental expression in a mouse model of Waardenburg-Shah (WS4) syndrome. Hum Mol Genet 15: 259–271 - PubMed
-
- Antonellis A, Huynh JL, Lee-Lin SQ, Vinton RM, Renaud G, Loftus SK, Elliot G, Wolfsberg TG, Green ED, McCallion AS, et al. 2008. Identification of neural crest and glial enhancers at the mouse Sox10 locus through transgenesis in zebrafish. PLoS Genet 4: e1000174 doi: 10.1371/journal.pgen.1000174 - PMC - PubMed
-
- Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K 2007. High-resolution profiling of histone methylations in the human genome. Cell 129: 823–837 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous
