TBC1D16 is a Rab4A GTPase activating protein that regulates receptor recycling and EGF receptor signaling
- PMID: 23019362
- PMCID: PMC3465424
- DOI: 10.1073/pnas.1204540109
TBC1D16 is a Rab4A GTPase activating protein that regulates receptor recycling and EGF receptor signaling
Abstract
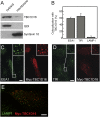
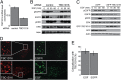
Rab4A is a master regulator of receptor recycling from endocytic compartments to the plasma membrane. The protein TBC1D16 is up-regulated in melanoma, and TBC1D16-overexpressing melanoma cells are dependent on TBC1D16. We show here that TBC1D16 enhances the intrinsic rate of GTP hydrolysis by Rab4A. TBC1D16 is both cytosolic and membrane associated; the membrane-associated pool colocalizes with transferrin and EGF receptors (EGFRs) and early endosome antigen 1, but not with LAMP1 protein. Expression of two TBC1D16 isoforms, but not the inactive R494A mutant, reduces transferrin receptor recycling but has no effect on transferrin receptor internalization. Expression of TBC1D16 alters GFP-Rab4A membrane localization. In HeLa cells, overexpression of TBC1D16 enhances EGF-stimulated EGFR degradation, concomitant with decreased EGFR levels and signaling. Thus, TBC1D16 is a GTPase activating protein for Rab4A that regulates transferrin receptor recycling and EGFR trafficking and signaling.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Stenmark H. Rab GTPases as coordinators of vesicle traffic. Nat Rev Mol Cell Biol. 2009;10:513–525. - PubMed
-
- Nguyen UT, et al. Analysis of the eukaryotic prenylome by isoprenoid affinity tagging. Nat Chem Biol. 2009;5:227–235. - PubMed
-
- Fukuda M. TBC proteins: GAPs for mammalian small GTPase Rab? Biosci Rep. 2011;31:159–168. - PubMed
-
- Pan X, Eathiraj S, Munson M, Lambright DG. TBC-domain GAPs for Rab GTPases accelerate GTP hydrolysis by a dual-finger mechanism. Nature. 2006;442:303–306. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

