Position-dependent correlations between DNA methylation and the evolutionary rates of mammalian coding exons
- PMID: 23019368
- PMCID: PMC3465446
- DOI: 10.1073/pnas.1208214109
Position-dependent correlations between DNA methylation and the evolutionary rates of mammalian coding exons
Abstract
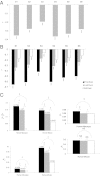
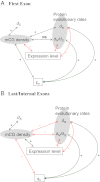
DNA cytosine methylation is a central epigenetic marker that is usually mutagenic and may increase the level of sequence divergence. However, methylated genes have been reported to evolve more slowly than unmethylated genes. Hence, there is a controversy on whether DNA methylation is correlated with increased or decreased protein evolutionary rates. We hypothesize that this controversy has resulted from the differential correlations between DNA methylation and the evolutionary rates of coding exons in different genic positions. To test this hypothesis, we compare human-mouse and human-macaque exonic evolutionary rates against experimentally determined single-base resolution DNA methylation data derived from multiple human cell types. We show that DNA methylation is significantly related to within-gene variations in evolutionary rates. First, DNA methylation level is more strongly correlated with C-to-T mutations at CpG dinucleotides in the first coding exons than in the internal and last exons, although it is positively correlated with the synonymous substitution rate in all exon positions. Second, for the first exons, DNA methylation level is negatively correlated with exonic expression level, but positively correlated with both nonsynonymous substitution rate and the sample specificity of DNA methylation level. For the internal and last exons, however, we observe the opposite correlations. Our results imply that DNA methylation level is differentially correlated with the biological (and evolutionary) features of coding exons in different genic positions. The first exons appear more prone to the mutagenic effects, whereas the other exons are more influenced by the regulatory effects of DNA methylation.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Reik W, Dean W, Walter J. Epigenetic reprogramming in mammalian development. Science. 2001;293:1089–1093. - PubMed
-
- Li E, Beard C, Jaenisch R. Role for DNA methylation in genomic imprinting. Nature. 1993;366:362–365. - PubMed
-
- Heard E, Clerc P, Avner P. X-chromosome inactivation in mammals. Annu Rev Genet. 1997;31:571–610. - PubMed
-
- Walsh CP, Chaillet JR, Bestor TH. Transcription of IAP endogenous retroviruses is constrained by cytosine methylation. Nat Genet. 1998;20:116–117. - PubMed
-
- Feinberg AP, Tycko B. The history of cancer epigenetics. Nat Rev Cancer. 2004;4:143–153. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

