Exploring chemical space for drug discovery using the chemical universe database
- PMID: 23019491
- PMCID: PMC3447393
- DOI: 10.1021/cn3000422
Exploring chemical space for drug discovery using the chemical universe database
Abstract
Herein we review our recent efforts in searching for bioactive ligands by enumeration and virtual screening of the unknown chemical space of small molecules. Enumeration from first principles shows that almost all small molecules (>99.9%) have never been synthesized and are still available to be prepared and tested. We discuss open access sources of molecules, the classification and representation of chemical space using molecular quantum numbers (MQN), its exhaustive enumeration in form of the chemical universe generated databases (GDB), and examples of using these databases for prospective drug discovery. MQN-searchable GDB, PubChem, and DrugBank are freely accessible at www.gdb.unibe.ch.
Figures


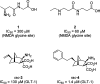
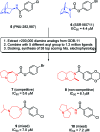

References
-
- van der Horst E.; Peironcely J. E.; van Westen G. J.; van den Hoven O. O.; Galloway W. R.; Spring D. R.; Wegner J. K.; van Vlijmen H. W.; Ijzerman A. P.; Overington J. P.; Bender A. (2011) Chemogenomics approaches for receptor deorphanization and extensions of the chemogenomics concept to phenotypic space. Curr. Top. Med. Chem. 11, 1964–1977. - PubMed
- Bon R. S.; Waldmann H. (2010) Bioactivity-guided navigation of chemical space. Acc. Chem. Res. 43, 1103–1114. - PubMed
-
- Harrison P. M.; Kumar A.; Lang N.; Snyder M.; Gerstein M. (2002) A question of size: the eukaryotic proteome and the problems in defining it. Nucleic Acids Res. 30, 1083–1090. - PMC - PubMed
- Montelione G. T.; Szyperski T. (2010) Advances in protein NMR provided by the NIGMS Protein Structure Initiative: impact on drug discovery. Curr. Opin. Drug Discovery Dev. 13, 335–349. - PMC - PubMed
- Bruckner A.; Polge C.; Lentze N.; Auerbach D.; Schlattner U. (2009) Yeast two-hybrid, a powerful tool for systems biology. Int. J. Mol. Sci. 10, 2763–2788. - PMC - PubMed
-
- Bleicher K. H.; Bohm H. J.; Muller K.; Alanine A. I. (2003) Hit and lead generation: Beyond high-throughput screening. Nat. Rev. Drug Discovery 2, 369–378. - PubMed
-
- Schuffenhauer A.; Brown N.; Selzer P.; Ertl P.; Jacoby E. (2005) Relationships between Molecular Complexity, Biological Activity, and Structural Diversity. J. Chem. Inf. Model. 46, 525–535. - PubMed
- Klebe G. (2006) Virtual ligand screening: strategies, perspectives and limitations. Drug Discovery Today 11, 580–594. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

