A comparative analysis of the intestinal metagenomes present in guinea pigs (Cavia porcellus) and humans (Homo sapiens)
- PMID: 23020652
- PMCID: PMC3472315
- DOI: 10.1186/1471-2164-13-514
A comparative analysis of the intestinal metagenomes present in guinea pigs (Cavia porcellus) and humans (Homo sapiens)
Abstract
Background: Guinea pig (Cavia porcellus) is an important model for human intestinal research. We have characterized the faecal microbiota of 60 guinea pigs using Illumina shotgun metagenomics, and used this data to compile a gene catalogue of its prevalent microbiota. Subsequently, we compared the guinea pig microbiome to existing human gut metagenome data from the MetaHIT project.
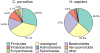
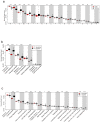
Results: We found that the bacterial richness obtained for human samples was lower than for guinea pig samples. The intestinal microbiotas of both species were dominated by the two phyla Bacteroidetes and Firmicutes, but at genus level, the majority of identified genera (320 of 376) were differently abundant in the two hosts. For example, the guinea pig contained considerably more of the mucin-degrading Akkermansia, as well as of the methanogenic archaea Methanobrevibacter than found in humans. Most microbiome functional categories were less abundant in guinea pigs than in humans. Exceptions included functional categories possibly reflecting dehydration/rehydration stress in the guinea pig intestine. Finally, we showed that microbiological databases have serious anthropocentric biases, which impacts model organism research.
Conclusions: The results lay the foundation for future gastrointestinal research applying guinea pigs as models for humans.
Figures

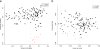
References
-
- Blaut M. Ecology and Physiology of the Intestinal Tract. Curr Top Microbiol Immunol. 2011. E-pub ahead of print. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

