Interferon regulatory factor 8/interferon consensus sequence binding protein is a critical transcription factor for the physiological phenotype of microglia
- PMID: 23020843
- PMCID: PMC3546867
- DOI: 10.1186/1742-2094-9-227
Interferon regulatory factor 8/interferon consensus sequence binding protein is a critical transcription factor for the physiological phenotype of microglia
Abstract
Background: Recent fate-mapping studies establish that microglia, the resident mononuclear phagocytes of the CNS, are distinct in origin from the bone marrow-derived myeloid lineage. Interferon regulatory factor 8 (IRF8, also known as interferon consensus sequence binding protein) plays essential roles in development and function of the bone marrow-derived myeloid lineage. However, little is known about its roles in microglia.
Methods: The CNS tissues of IRF8-deficient mice were immunohistochemically analyzed. Pure microglia isolated from wild-type and IRF8-deficient mice were studied in vitro by proliferation, immunocytochemical and phagocytosis assays. Microglial response in vivo was compared between wild-type and IRF8-deficient mice in the cuprizon-induced demyelination model.
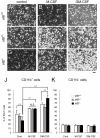
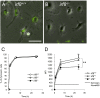
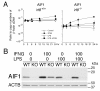
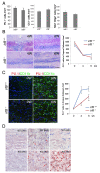
Results: Our analysis of IRF8-deficient mice revealed that, in contrast to compromised development of IRF8-deficient bone marrow myeloid lineage cells, development and colonization of microglia are not obviously affected by loss of IRF8. However, IRF8-deficient microglia demonstrate several defective phenotypes. In vivo, IRF8-deficient microglia have fewer elaborated processes with reduced expression of IBA1/AIF1 compared with wild-type microglia, suggesting a defective phenotype. IRF8-deficient microglia are significantly less proliferative in mixed glial cultures than wild-type microglia. Unlike IRF8-deficient bone marrow myeloid progenitors, exogenous macrophage colony stimulating factor (colony stimulating factor 1) (M-CSF (CSF1)) restores their proliferation in mixed glial cultures. In addition, IRF8-deficient microglia exhibit an exaggerated growth response to exogenous granulocyte-macrophage colony stimulating factor (colony stimulating factor 2) (GM-CSF (CSF2)) in the presence of other glial cells. IRF8-deficient microglia also demonstrate altered cytokine expressions in response to interferon-gamma and lipopolysaccharide in vitro. Moreover, the maximum phagocytic capacity of IRF8-deficient microglia is reduced, although their engulfment of zymosan particles is not overtly impaired. Defective scavenging activity of IRF8-deficient microglia was further confirmed in vivo in the cuprizone-induced demyelination model in mice.
Conclusions: This study is the first to demonstrate the essential contribution of IRF8-mediated transcription to a broad range of microglial phenotype. Microglia are distinct from the bone marrow myeloid lineage with respect to their dependence on IRF8-mediated transcription.
Figures





References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

