Protein modularity, cooperative binding, and hybrid regulatory states underlie transcriptional network diversification
- PMID: 23021217
- PMCID: PMC3519278
- DOI: 10.1016/j.cell.2012.08.018
Protein modularity, cooperative binding, and hybrid regulatory states underlie transcriptional network diversification
Abstract
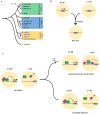
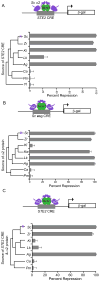
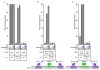
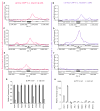
We examine how different transcriptional network structures can evolve from an ancestral network. By characterizing how the ancestral mode of gene regulation for genes specific to a-type cells in yeast species evolved from an activating paradigm to a repressing one, we show that regulatory protein modularity, conversion of one cis-regulatory sequence to another, distribution of binding energy among protein-protein and protein-DNA interactions, and exploitation of ancestral network features all contribute to the evolution of a novel regulatory mode. The formation of this derived mode of regulation did not disrupt the ancestral mode and thereby created a hybrid regulatory state where both means of transcription regulation (ancestral and derived) contribute to the conserved expression pattern of the network. Finally, we show how this hybrid regulatory state has resolved in different ways in different lineages to generate the diversity of regulatory network structures observed in modern species.
Copyright © 2012 Elsevier Inc. All rights reserved.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

