miR-211 is a prosurvival microRNA that regulates chop expression in a PERK-dependent manner
- PMID: 23022383
- PMCID: PMC3496065
- DOI: 10.1016/j.molcel.2012.08.025
miR-211 is a prosurvival microRNA that regulates chop expression in a PERK-dependent manner
Abstract
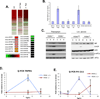
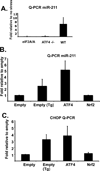
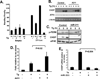
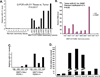
MicroRNAs typically function at the level of posttranscriptional gene silencing within the cytoplasm; however, increasing evidence suggests that they may also function in nuclear, Argonaut-containing complexes, to directly repress target gene transcription. We have investigated the role of microRNAs in mediating endoplasmic reticulum (ER) stress responses. ER stress triggers the activation of three signaling molecules: Ire-1α/β, PERK, and ATF6, whose function is to facilitate adaption to the ensuing stress. We demonstrate that PERK induces miR-211, which in turn attenuates stress-dependent expression of the proapoptotic transcription factor chop/gadd153. MiR-211 directly targets the proximal chop/gadd153 promoter, where it increases histone methylation and represses chop expression. Maximal chop accumulation ultimately correlates with miR-211 downregulation. Our data suggest a model in which PERK-dependent miR-211 induction prevents premature chop accumulation and thereby provides a window of opportunity for the cell to re-establish homeostasis prior to apoptotic commitment.
Copyright © 2012 Elsevier Inc. All rights reserved.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

