Systems-level characterization of a host-microbe metabolic symbiosis in the mammalian gut
- PMID: 23022739
- PMCID: PMC3555882
- DOI: 10.4161/gmic.22370
Systems-level characterization of a host-microbe metabolic symbiosis in the mammalian gut
Abstract
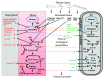
The human gut microbiota consists of ten times more microorganisms than there are cells in our body, processes otherwise indigestible nutrients, and produces important energy precursors, essential amino acids, and vitamins. In this study, we assembled and validated a genome-scale metabolic reconstruction of Bacteroides thetaiotaomicron (iAH991), a prominent representative of the human gut microbiota, consisting of 1488 reactions, 1152 metabolites, and 991 genes. To create a comprehensive metabolic model of host-microbe interactions, we integrated iAH991 with a previously published mouse metabolic reconstruction, which was extended for intestinal transport and absorption reactions. The two metabolic models were linked through a joint compartment, the lumen, allowing metabolite exchange and providing a route for simulating different dietary regimes. The resulting model consists of 7239 reactions, 5164 metabolites, and 2769 genes. We simultaneously modeled growth of mouse and B. thetaiotaomicron on five different diets varying in fat, carbohydrate, and protein content. The integrated model captured mutually beneficial cross-feeding as well as competitive interactions. Furthermore, we identified metabolites that were exchanged between the two organisms, which were compared with published metabolomics data. This analysis resulted for the first time in a comprehensive description of the co-metabolism between a host and its commensal microbe. We also demonstrate in silico that the presence of B. thetaiotaomicron could rescue the growth phenotype of the host with an otherwise lethal enzymopathy and vice versa. This systems approach represents a powerful tool for modeling metabolic interactions between a gut microbe and its host in health and disease.
Keywords: Bacteroidesthetaiotaomicron; Mus musculus; computational modeling; constraint-based modeling; host-microbe interactions; metabolism; systems biology.
Figures

References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
