Cell type and context-specific function of PLAG1 for IGF2 P3 promoter activity
- PMID: 23023303
- PMCID: PMC3583874
- DOI: 10.3892/ijo.2012.1641
Cell type and context-specific function of PLAG1 for IGF2 P3 promoter activity
Abstract
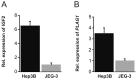
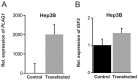
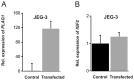
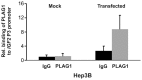
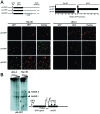
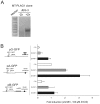
The fetal transcription factor PLAG1 is found to be overexpressed in cancers, and has been suggested to bind the insulin like growth factor 2 (IGF2) P3 promoter, and to activate the IGF2 gene. The expression of IGF2 has partly been linked to loss of CTCF-dependent chromatin insulator function at the H19 imprinting control region (ICR). We investigated the role of PLAG1 for IGF2 regulation in Hep3B and JEG-3 cell lines. Chromatin immunoprecipitation revealed cell type-specific binding of PLAG1 to the IGF2 P3 promoter, which was substantially insensitive to recombinant PLAG1 overexpression in the endogenous context. We hypothesized that the H19 chromatin insulator may be involved in the cell type-specific PLAG1 response. By using a GFP reporter gene/insulator assay plasmid construct with and without the H19 ICR and/or an SV40 enhancer, we confirm that the effect of the insulator is specifically associated with the activity of the IGF2 P3 promoter in the GFP reporter system, and furthermore, that the reporter insulator is functional in JEG-3 but not in Hep3B cells. FACS analysis was used to assess the function of PLAG1 in low endogenously expressing, but Zn-inducible stable PLAG1 expressing JEG-3 cell clones. Considerable increase in IGF2 expression upon PLAG1 induction with a partial insulator overriding activity was found using the reporter constructs. This is in contrast to the effect of the endogenous IGF2 gene which was insensitive to PLAG1 expression in JEG-3, while modestly induced the already highly expressed IGF2 gene in Hep3B cells. We suggest that the PLAG1 binding to the IGF2 P3 promoter and IGF2 expression is cell type-specific, and that the PLAG1 transcription factor acts as a transcriptional facilitator that partially overrides the insulation by the H19 ICR.
Figures
References
-
- LeRoith D, Roberts CT., Jr The insulin-like growth factor system and cancer. Cancer Lett. 2003;195:127–137. - PubMed
-
- Cui H, Cruz-Correa M, Giardiello FM, et al. Loss of IGF2 imprinting: a potential marker of colorectal cancer risk. Science. 2003;299:1753–1755. - PubMed
-
- Feinberg AP, Cui H, Ohlsson R. DNA methylation and genomic imprinting: insights from cancer into epigenetic mechanisms. Semin Cancer Biol. 2002;12:389–398. - PubMed
-
- Ohlsson R, Kanduri C, Whitehead J, Pfeifer S, Lobanenkov V, Feinberg AP. Epigenetic variability and the evolution of human cancer. Adv Cancer Res. 2003;88:145–168. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

