The molecular basis of T cell acute lymphoblastic leukemia
- PMID: 23023710
- PMCID: PMC3461904
- DOI: 10.1172/JCI61269
The molecular basis of T cell acute lymphoblastic leukemia
Abstract
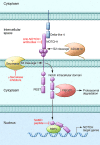
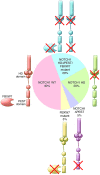
T cell acute lymphoblastic leukemias (T-ALLs) arise from the malignant transformation of hematopoietic progenitors primed toward T cell development, as result of a multistep oncogenic process involving constitutive activation of NOTCH signaling and genetic alterations in transcription factors, signaling oncogenes, and tumor suppressors. Notably, these genetic alterations define distinct molecular groups of T-ALL with specific gene expression signatures and clinicobiological features. This review summarizes recent advances in our understanding of the molecular genetics of T-ALL.
Figures
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

