ChIP-exo method for identifying genomic location of DNA-binding proteins with near-single-nucleotide accuracy
- PMID: 23026909
- PMCID: PMC3813302
- DOI: 10.1002/0471142727.mb2124s100
ChIP-exo method for identifying genomic location of DNA-binding proteins with near-single-nucleotide accuracy
Abstract
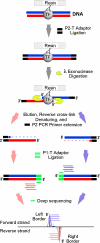
This unit describes the ChIP-exo methodology, which combines chromatin immunoprecipitation (ChIP) with lambda exonuclease digestion followed by high-throughput sequencing. ChIP-exo allows identification of a nearly complete set of the binding locations of DNA-binding proteins at near-single-nucleotide resolution with almost no background. The process is initiated by cross-linking DNA and associated proteins. Chromatin is then isolated from nuclei and subjected to sonication. Subsequently, an antibody against the desired protein is used to immunoprecipitate specific DNA-protein complexes. ChIP DNA is purified, sequencing adaptors are ligated, and the adaptor-ligated DNA is then digested by lambda exonuclease, generating 25- to 50-nucleotide fragments for high-throughput sequencing. The sequences of the fragments are mapped back to the reference genome to determine the binding locations. The 5' ends of DNA fragments on the forward and reverse strands indicate the left and right boundaries of the DNA-protein binding regions, respectively.
2012 by John Wiley & Sons, Inc.
Figures
References
Literature Cited
-
- Lee TI, Rinaldi NJ, Robert F, Odom DT, Bar-Joseph Z, Gerber GK, Hannett NM, Harbison CT, Thompson CM, Simon I, et al. Transcriptional regulatory networks in Saccharomyces cerevisiae. Science. 2002;298:799–804. - PubMed
-
- Little JW. Lambda exonuclease. Gene Amplif Anal. 1981;2:135–145. - PubMed
-
- Ptashne M, Gann A. Transcriptional activation by recruitment. Nature. 1997;386:569–577. - PubMed
Key Reference
-
- Rhee HS, Pugh BF. This is the first paper, which described a ChIP-exo method for various transcription factors in yeast to human. Moreover, this paper shows the proof of principle of ChIP-exo, which identified a nearly complete set of genomic bound locations at near nucleotide resolution. 2011
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

